KMT2A Antibody
-
货号:CSB-PA612015
-
规格:¥880
-
图片:
-
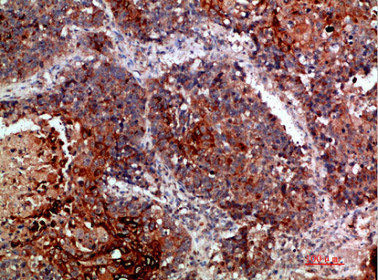
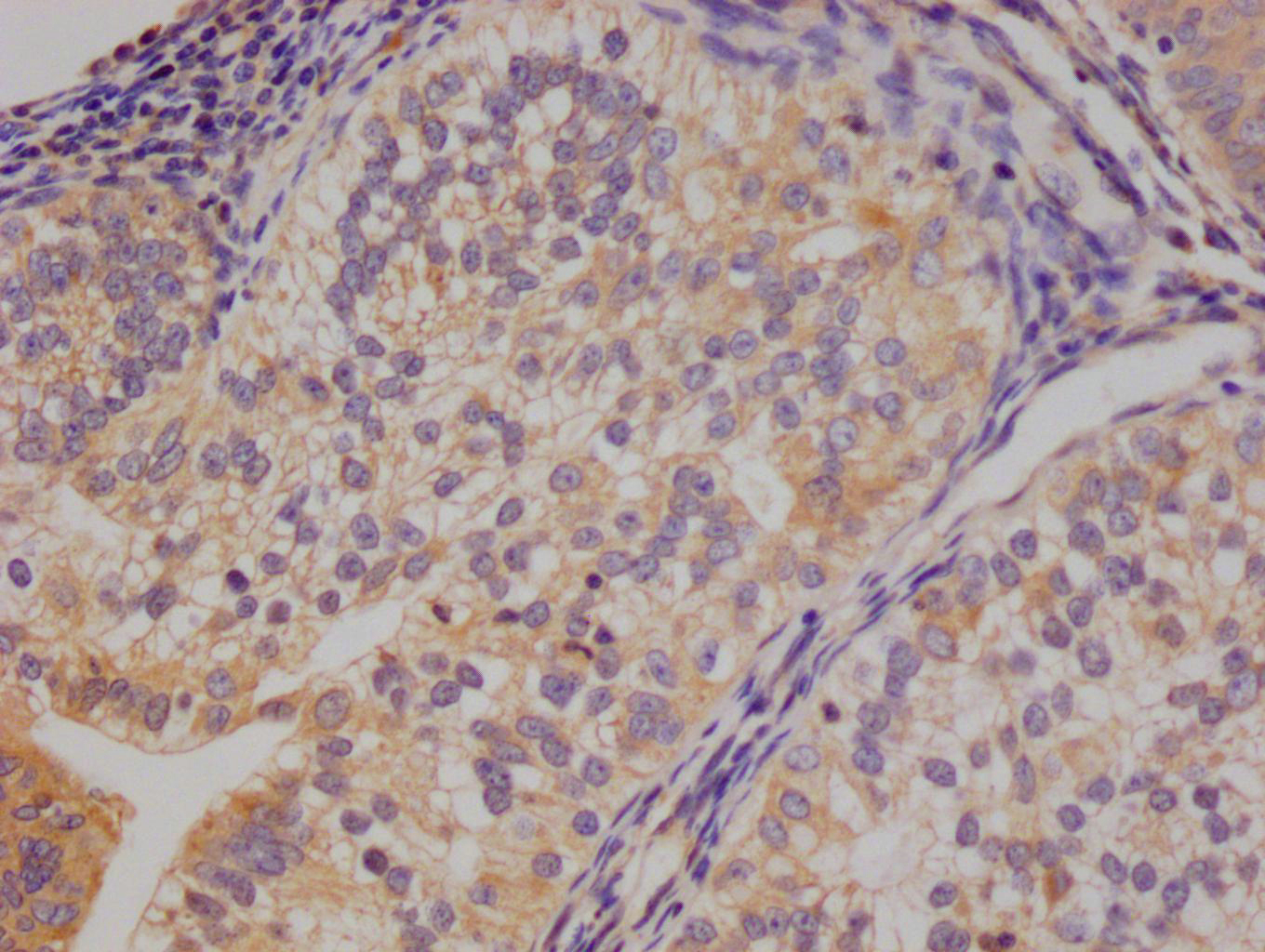
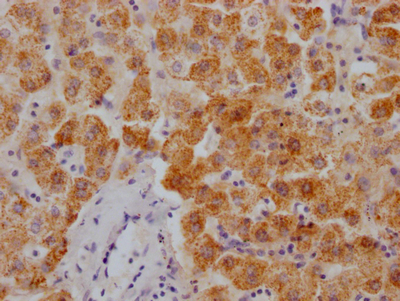
Immunohistochemical analysis of paraffin-embedded human-lung-cancer, antibody was diluted at 1:200
-
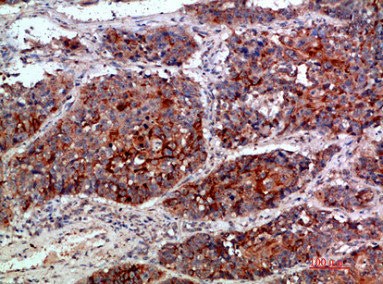
Immunohistochemical analysis of paraffin-embedded human-lung-cancer, antibody was diluted at 1:200
-
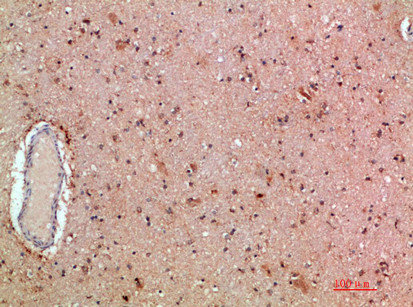
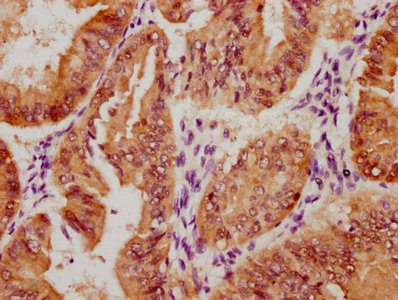
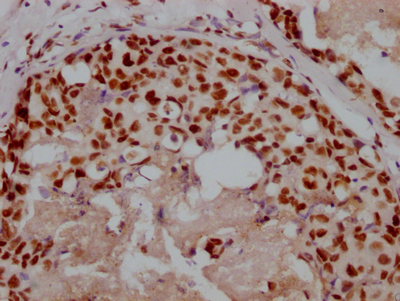
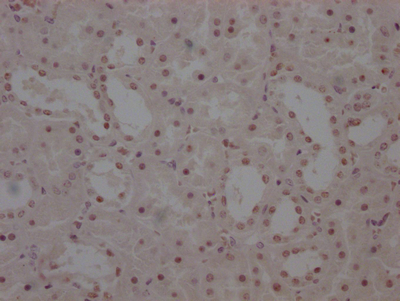
Immunohistochemical analysis of paraffin-embedded human-brain, antibody was diluted at 1:200
-
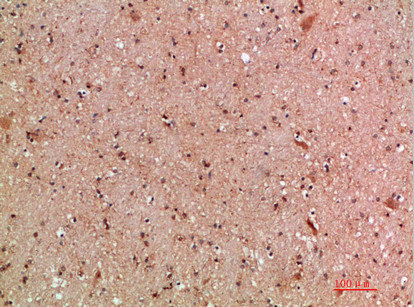
Immunohistochemical analysis of paraffin-embedded human-brain, antibody was diluted at 1:200
-
-
其他:
产品详情
-
Uniprot No.:Q03164
-
基因名:
-
别名:ALL-1 antibody; ALL1 antibody; C-terminal cleavage product of 180 kDa antibody; CXXC-type zinc finger protein 7 antibody; CXXC7 antibody; HRX antibody; HTRX1 antibody; KMT2A antibody; Lysine N-methyltransferase 2A antibody; Mll antibody; MLL cleavage product C180 antibody; MLL1 antibody; MLL1_HUMAN antibody; MLL1A antibody; N-terminal cleavage product of 320 kDa antibody; p180 antibody; p320 antibody; Trithorax-like protein antibody; TRX1 antibody; Zinc finger protein HRX antibody
-
宿主:Rabbit
-
反应种属:Human,Mouse,Rat
-
免疫原:Synthetic peptide from Human protein at AA range: 3850-3900
-
免疫原种属:Homo sapiens (Human)
-
标记方式:Non-conjugated
-
纯化方式:The antibody was affinity-purified from rabbit serum by affinity-chromatography using specific immunogen.
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:PBS, pH 7.4, containing 0.02% sodium azide as Preservative and 50% Glycerol.
-
产品提供形式:Liquid
-
应用范围:IHC,ELISA
-
推荐稀释比:
Application Recommended Dilution IHC IHC-p:1:50-300 ELISA 1:10000-20000 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
靶点详情
-
功能:Histone methyltransferase that plays an essential role in early development and hematopoiesis. Catalytic subunit of the MLL1/MLL complex, a multiprotein complex that mediates both methylation of 'Lys-4' of histone H3 (H3K4me) complex and acetylation of 'Lys-16' of histone H4 (H4K16ac). Catalyzes methyl group transfer from S-adenosyl-L-methionine to the epsilon-amino group of 'Lys-4' of histone H3 (H3K4) via a non-processive mechanism. Part of chromatin remodeling machinery predominantly forms H3K4me1 and H3K4me2 methylation marks at active chromatin sites where transcription and DNA repair take place. Has weak methyltransferase activity by itself, and requires other component of the MLL1/MLL complex to obtain full methyltransferase activity. Has no activity toward histone H3 phosphorylated on 'Thr-3', less activity toward H3 dimethylated on 'Arg-8' or 'Lys-9', while it has higher activity toward H3 acetylated on 'Lys-9'. Binds to unmethylated CpG elements in the promoter of target genes and helps maintain them in the nonmethylated state. Required for transcriptional activation of HOXA9. Promotes PPP1R15A-induced apoptosis. Plays a critical role in the control of circadian gene expression and is essential for the transcriptional activation mediated by the CLOCK-ARNTL/BMAL1 heterodimer. Establishes a permissive chromatin state for circadian transcription by mediating a rhythmic methylation of 'Lys-4' of histone H3 (H3K4me) and this histone modification directs the circadian acetylation at H3K9 and H3K14 allowing the recruitment of CLOCK-ARNTL/BMAL1 to chromatin. Also has auto-methylation activity on Cys-3882 in absence of histone H3 substrate.
-
基因功能参考文献:
- our work shows that ALOX5 plays a moderate anti-tumor role and functions as a drug sensitizer, with a therapeutic potential, in MLL-rearranged AML. PMID: 28500307
- Studied lysine methyltransferase 2A (KMT2A) genetic rearrangements in child and young adult T-lymphoblastic leukemia/lymphoma. PMID: 30203571
- Our experience suggests that AML with KMT2A rearrangement should be considered for the differential diagnosis of infantile cases with atypical monocytosis suggestive of JMML. Cytogenetic studies, including fluorescence in situ hybridization analysis of KMT2A, may be helpful in distinguishing between AML with KMT2A rearrangement and JMML. PMID: 30143999
- Replication stress-induced recruitment of EndoG to the MLLbcr. PMID: 28626219
- Wiedemann-Steiner syndrome causative splice and missense variants lead to reduction of KMT2A function. PMID: 29203834
- SETD2 is required to maintain high H3K79 di-methylation and MLL-AF9-binding to critical target genes, such as Hoxa9. PMID: 29777171
- One of the USP10 targets is TP53. Wildtype TP53 is usually rescued from proteasomal degradation by USP10. As most KMT2A leukemias display wildtype p53 alleles, one might argue that the disruption of an USP10 allele can be classified as a pro-oncogenic event. PMID: 30107050
- this work highlights that MLLr AML comprises of an AML subgroup affecting younger patients with poor prognosis. The allo-HSCT in MRD-negative patients could be the best therapeutic option. PMID: 29384595
- Myeloid Zinc Finger 1 and GA Binding Protein Co-Operate with Sox2 in Regulating the Expression of Yes-Associated Protein 1 in Cancer Cells PMID: 28905448
- This study demonstrates a tractable approach for respecifying iPSC-derived blood cells into highly engraftable hematopoietic stem and progenitor cells (HSPCs) through transient expression of a single transcription factor, MLL-AF4 PMID: 29386396
- the influence of TGFbeta/SMADs on MLL- and MLL-AF4-mediated 5-LO promoter activation PMID: 28803964
- Chromosomal translocations involving MLL1 appear to directly perturb the regulation of multiple chromatin-associated complexes to allow inappropriate expression of developmentally regulated genes and thus drive leukemia development. [review] PMID: 28242784
- FLT3 cell-surface expression did not vary by FLT3 mutational status, but high FLT3 expression was strongly associated with KMT2A rearrangements. Our study found that there was no prognostic significance of FLT3 cell surface expression in pediatric Acute Myeloid Leukemia PMID: 28108543
- results indicate that KMT2A promotes melanoma growth by activating the hTERT signaling, suggesting that the KMT2A/hTERT signaling pathway may be a potential therapeutic target for melanoma. PMID: 28726783
- A novel regulatory pathway for MLL1, which may open a new therapeutic approach to MLL-rearrangement leukaemia. PMID: 28939057
- data confirm MLL-PTD and, to a lesser extent, FLT3-ITD as common events in +11 AML.6, 7, 8 However, the high mutation frequencies of U2AF1 and genes involved in methylation (DNMT3A, IDH2) have hitherto not been reported in +11 AML PMID: 27435003
- It is, to our knowledge, the first case of B acute lymphoblastic leukemia with this fusion gene. At the molecular level, two rearrangements were detected using RNA sequencing juxtaposing exon 7 to exon 2 and exon 9 to intron 1-2 of the KMT2A and MAML2 genes respectively, and one rearrangement using Sanger sequencing juxtaposing exon 8 and exon 2. PMID: 28535805
- The most common KMT2A breakpoint region was intron/exon 9 (3/8 patients), followed by intron/exon 11 and 10. PMID: 27282883
- study describes 2 patients with Wiedemann-Steiner syndrome who presented with variable severity; findings revealed a de novo nonsense mutation, p.Gln1978*, of KMT2A in the former, and a missense mutation, p.Gly1168Asp, in the latter PMID: 27777327
- Comprehensive genetic analysis of donor cell derived leukemia with KMT2A rearrangement PMID: 28921816
- Data show that expression of myeloid-lymphoid leukemia protein (MLL) fusion protein does neither influence DNA signaling nor DNA double strand breaks (DNA-DSBs) repair. PMID: 27119507
- In this case study, we diagnosed t-MN with KMT2A rearrangement in a patient with history of B-ALL with 9p deletion and gain of X chromosome. Unusual features associated with this case are discussed PMID: 29155023
- Identification of novel biomarkers for MLL-translocated acute myeloid leukemia. PMID: 28911906
- Collectively, these data indicated that ATR or ATM inhibition represent potential therapeutic strategies for the treatment of AML, especially MLL-driven leukemias. PMID: 27625305
- different cancer mutations in MLL1 lead to a loss or increase in activity, illustrating the complex and tumor-specific role of MLL1 in carcinogenesis. PMID: 28182322
- we report two boys with novel KMT2A mutations from Chinese origin for the first time. They do not show one of the characteristic WDSTS phenotype, cubiti hypertrichosis. Instead, both of them had absent palmar proximal transverse crease. The feature was not linked to WDSTS patients previously. Our findings extend the WDSTS phenotypic spectrum. PMID: 27759909
- Whole exome sequencing allowed identifying a previously unreported de novo KMT2A missense mutation affecting the DNA binding domain of the methyltransferase. This finding expands the clinical phenotype associated with KMT2A mutations to include immunodeficiency and epilepsy as clinically relevant features for this disorder. PMID: 27320412
- These results define an important role for MLL1 in regulating macrophage-mediated inflammation in wound repair and identify a potential target for the treatment of chronic inflammation in diabetic wounds. PMID: 28663191
- MLL rearrangement is associated with infant acute lymphoblastic leukemia. PMID: 27588400
- These results reveal a cooperative transcriptional activation mechanism of AEP and DOT1L and suggest a molecular rationale for the simultaneous inhibition of the MLL fusion-AF4 complex and DOT1L for more effective treatment of MLL-rearranged leukemia. PMID: 28394257
- this indicates that DOT1L function, like MLL, does not completely rely on its methyltransferase activity. Nevertheless, the small molecule DOT1L inhibition is sufficient to block the proliferation of MLL fusion-induced leukemia cells of murine and human origin PMID: 26923329
- Studies demonstrate that histone methyl-transferase MLL1 coordinates with HIF1alpha and histone acetyltransferase p300 and regulate hypoxia-induced HOTAIR expression. The hypoxia-induced upregulation of HOTAIR expression may contribute to its roles in tumorigenesis. PMID: 28756022
- MLL1 and MLL2 collaborate to regulate gene expression and leukemia maintenance not through redundancy, but through distinct pathways. PMID: 28609655
- MLL1, deposits H3K4me2 at CpG-dense regions that could serve as polycomb response elements. PMID: 27447986
- A patient was diagnosed with Wiedemann-Steiner syndrome based on identification of a novel de novo pathogenic variant in the KMT2A gene. PMID: 28359930
- MLL gene rearrangement is associated with acute lymphocytic leukemia. PMID: 27343252
- Our data thus extend MLL1 function to H3K4 methylation and PD-L1 transcription activation in pancreatic cancer cells. PMID: 28131992
- KMT2A gene rearrangement is associated with acute megakaryoblastic leukemia in non-Down syndrome. PMID: 28063190
- The data revealed the presence of a partial tandem duplication of the MLL gene as the only detectable copy number change and 11 non-silent somatic mutations, including DNMT3A R882H and NRAS G13D. All somatic lesions were present both at initial MDS/MPN-U diagnosis and at AML presentation at similar mutant allele frequencies. PMID: 27240832
- KMT2A rearrangement is associated with B-cell lymphoblastic leukemia. PMID: 27786413
- Loss of MLL-AF4 activity results in a reduction of H3K79me3 levels in the gene body and H3K27Ac levels at the 3' BCL-2 enhancer, revealing a novel regulatory link between these two histone marks and MLL-AF4-mediated activation of BCL-2. PMID: 27856324
- Results revealed that double minute chromosomes in myeloid neoplasms commonly harbor MYC or MLL gene amplification and manifest as micronuclei within leukemic blasts. PMID: 27318442
- Targeted Disruption of the Interaction between WD-40 Repeat Protein 5 (WDR5) and Mixed Lineage Leukemia (MLL)/SET1 Family Proteins Specifically Inhibits MLL1 and SETd1A Methyltransferase Complexes. PMID: 27563068
- miR-155 levels are correlated with MLL translocations, but that miR-155 expression is dispensable for the formation of AML and has no effect on leukemia progression. PMID: 27619068
- MLL-ENL dysregulated the proliferative and repopulating capacity of hematopoietic stem cells. PMID: 25456127
- This review summarizes and discusses the general mechanisms of H3K4 methylation, and how the six main enzymes from the SET/MLL family (responsible for H3K4me1/me2/me3) function in hematopoiesis and in hematologic malignancies. [review] PMID: 27796741
- Results show that human MLL-AF9 expression in mouse long-term hematopoietic stem cells causes invasive, chemoresistant acute myeloid leukemia that expresses genes related to epithelial-mesenchymal transition. PMID: 27344946
- s evaluated the expression levels of the MLL gene in AML patients. PMID: 27561414
- Results show that MLL-AF9 reduces Id2 and increases E2-2 expression to drive and sustain leukemia stem cell potential in MLL-rearranged acute myeloid leukemia (AML). Low expression of Id2 or of an Id2 gene signature is associated with poor prognosis in not only MLL-rearranged but also t(8;21) AML patients. PMID: 27374225
- Pygo2 functions as a prognostic factor for glioma due to its up-regulation of H3K4me3 and promotion of MLL1/MLL2 complex recruitment. PMID: 26902498
显示更多
收起更多
-
相关疾病:Wiedemann-Steiner syndrome (WDSTS)
-
亚细胞定位:Nucleus.; [MLL cleavage product N320]: Nucleus.; [MLL cleavage product C180]: Nucleus. Note=Localizes to a diffuse nuclear pattern when not associated with MLL cleavage product N320.
-
蛋白家族:Class V-like SAM-binding methyltransferase superfamily, Histone-lysine methyltransferase family, TRX/MLL subfamily
-
组织特异性:Heart, lung, brain and T- and B-lymphocytes.
-
数据库链接:
HGNC: 7132
OMIM: 159555
KEGG: hsa:4297
STRING: 9606.ENSP00000436786
UniGene: Hs.258855
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-