NRAS Antibody
-
货号:CSB-PA016070LA01HU
-
规格:¥440
-
促销:
-
图片:
-
其他:
产品详情
-
产品名称:Rabbit anti-Homo sapiens (Human) NRAS Polyclonal antibody
-
Uniprot No.:P01111
-
基因名:
-
别名:ALPS4 antibody; AV095280 antibody; GTPase NRas antibody; HRAS1 antibody; N ras antibody; N ras protein part 4 antibody; Neuroblastoma RAS viral (v ras) oncogene homolog antibody; NRAS antibody; NRAS1 antibody; NS6 antibody; OTTHUMP00000013879 antibody; OTTMUSP00000023521 antibody; RASN_HUMAN antibody; Transforming protein N Ras antibody; Transforming protein N-Ras antibody; v ras neuroblastoma RAS viral oncogene homolog antibody
-
宿主:Rabbit
-
反应种属:Human
-
免疫原:Recombinant Human GTPase NRas protein (1-186AA)
-
免疫原种属:Homo sapiens (Human)
-
标记方式:Non-conjugated
本页面中的产品,NRAS Antibody (CSB-PA016070LA01HU),的标记方式是Non-conjugated。对于NRAS Antibody,我们还提供其他标记。见下表:
-
克隆类型:Polyclonal
-
抗体亚型:IgG
-
纯化方式:>95%, Protein G purified
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, pH 7.4 -
产品提供形式:Liquid
-
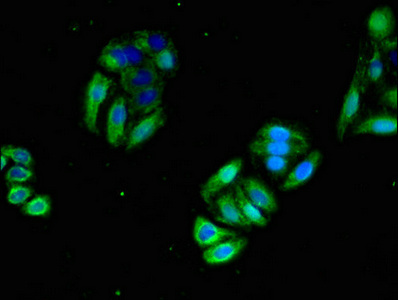
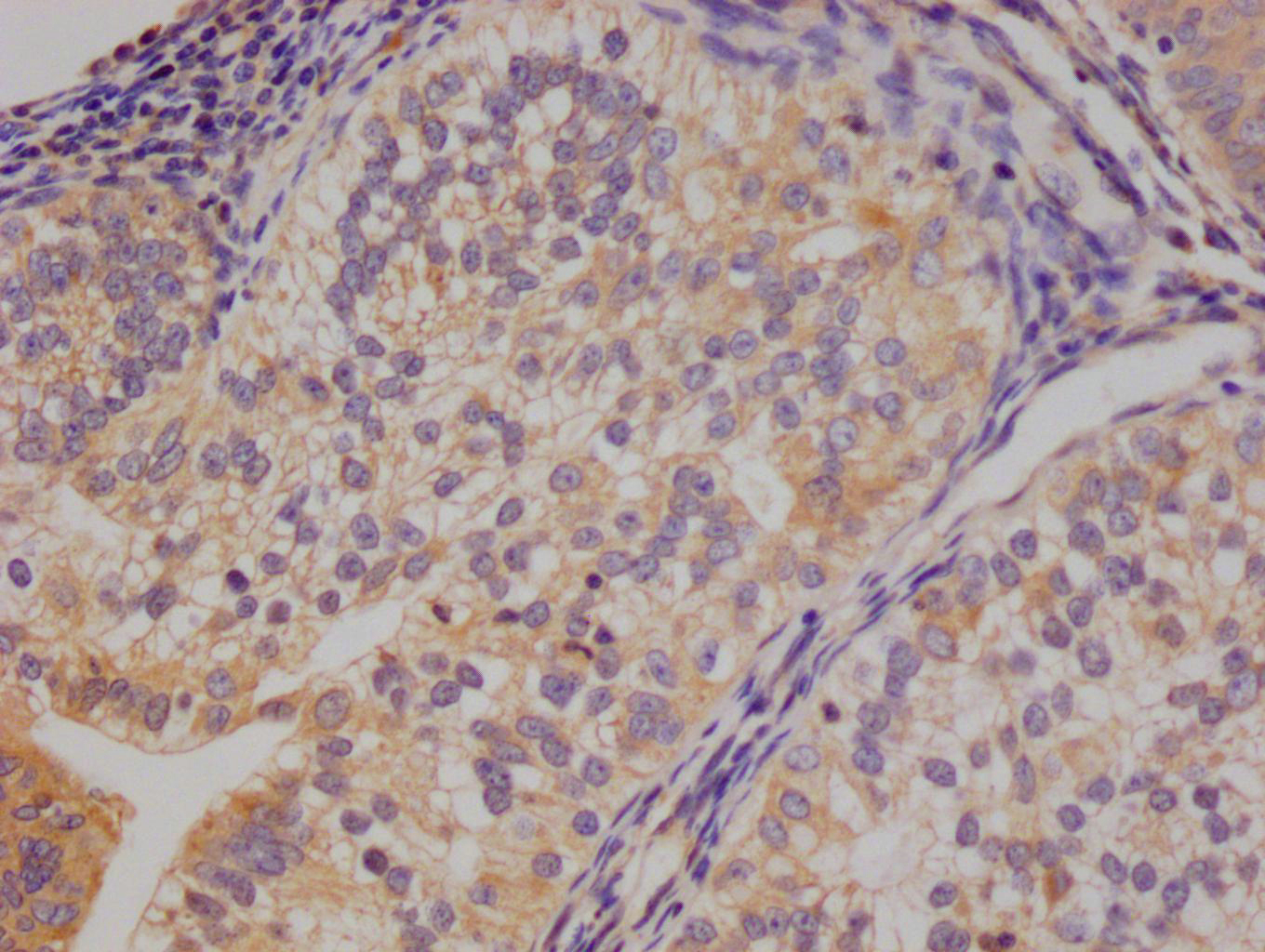
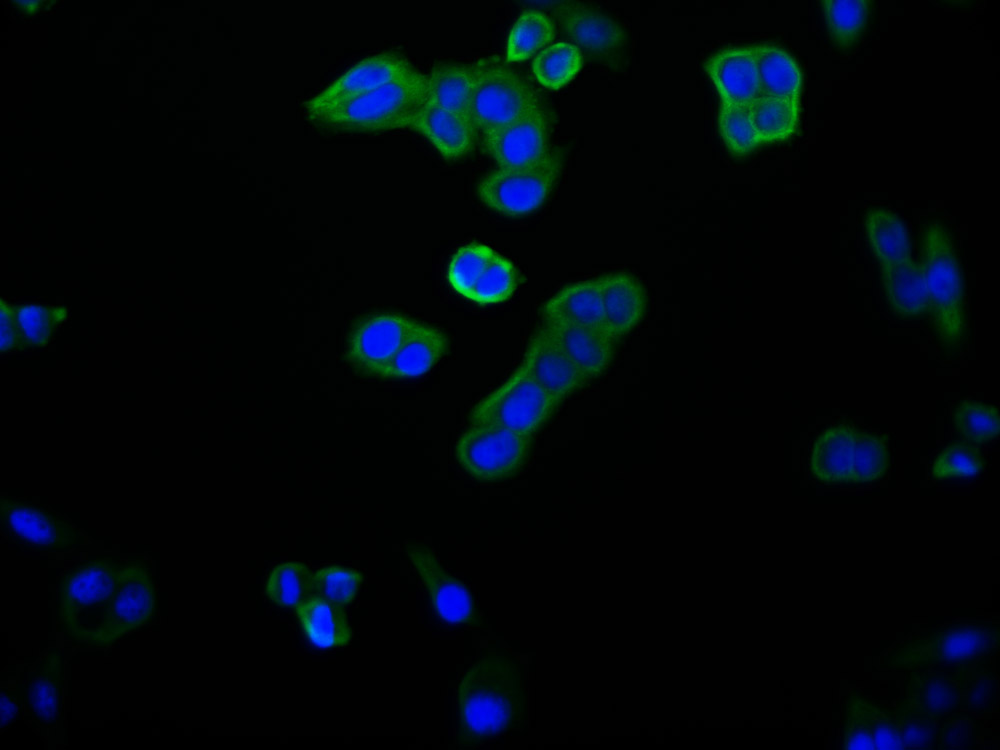
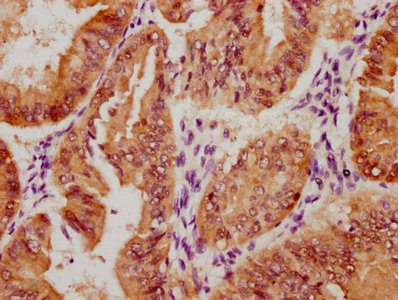
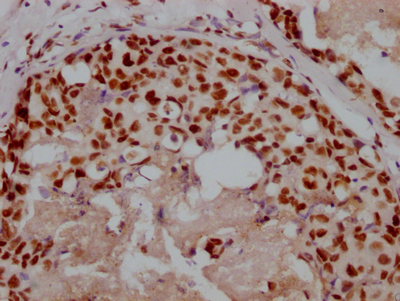
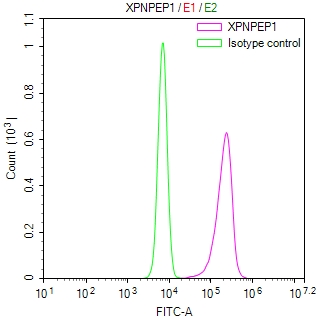
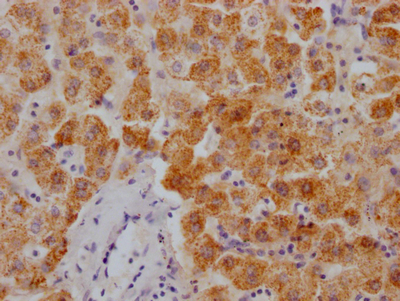
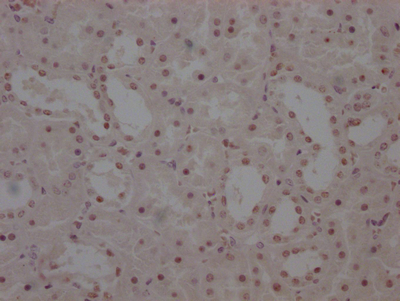
应用范围:ELISA, IHC, IF
-
推荐稀释比:
Application Recommended Dilution IHC 1:20-1:200 IF 1:50-1:200 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Ras proteins bind GDP/GTP and possess intrinsic GTPase activity.
-
基因功能参考文献:
- Tet2/Nras double-mutant leukemia showed preferential sensitivity to MAPK kinase (MEK) inhibition in both mouse model and patient samples PMID: 29275866
- Our study provided the first evidence that miR-29a suppressed lung cancer cell growth through inhibition of NRAS PMID: 29495918
- NRAS mutations harbor commonly in neurocutaneous melanosis. PMID: 30145692
- Ets-1 is induced by BRAF or MEK kinase inhibition, resulting in increased NRAS expression, which could be blocked by inactivation of Usp9x. PMID: 28198367
- we used hot-spot mutation sequencing to examine whether KRAS/NRAS mutations, a characteristic feature of mesonephric carcinoma,1 are also present in mesonephric hyperplasia. None of the mesonephric hyperplasia cases harboured a KRAS or NRAS mutation. PMID: 28703285
- BRAF mutations more frequently affected individuals younger than 61 with phototype II. In contrast, NRAS mutations were more frequent in phototype III cases. Mutations of both genes were more frequent in cases with satellitosis in the first melanoma, and in cases with ulceration in the subsequent lesions. PMID: 29180316
- Identification of KRAS/NRAS/BRAF mutation status is crucial to predict the therapeutic effect and determine individual therapeutic strategies for patients with colorectal cancer. PMID: 29335867
- common conjunctival melanocytic nevi have mutually exclusive mutations in BRAF and NRAS. The two conjunctival blue nevi harbored GNAQ mutations. This suggests the driver mutations of conjunctival nevi are similar to those of nevi of the skin. At the molecular level, conjunctival nevi appear more like cutaneous nevi than choroidal nevi PMID: 29332123
- the mutational status of BRAF, NRAS, and TERT promoter genes in 97 melanomas, was investigated. PMID: 29061376
- Deciphering KRAS and NRAS mutated clone dynamics in MLL-AF4 paediatric leukaemia by ultra deep sequencing analysis. PMID: 27698462
- N-Ras preferentially populated raft domains when bound to mant-GDP. It lost its preference for rafts when associated with a GTP mimic, mant-GppNHp. The isolated lipidated C-terminal peptide of N-Ras was outside of the liquid-ordered rafts, most likely in the bulk-disordered lipid. Substitution of the N-Ras N-terminal G domain with a homologous G domain of H-Ras disrupted the nucleotide-dependent lipid domain switch. PMID: 29280621
- We report herein for the first time that 30% of cutaneous NRAS mutant melanomas have a high M%NRAS. Chromosome instability, (chromosome 1 polysomy, intratumor copy number variation of chromosome1/NRAS) rather than the acquired copy neutral LOH seems to be responsible for most of the cases with high M%NRAS. PMID: 28668077
- NRAS status (exons 2-4) was analyzed by Pyrosequencing in a case series of 50 squamous cell anal carcinoma patients. PMID: 27886225
- NRAS-mutant tumors tended to behave more aggressively particularly in early stages of the disease in studied high-risk melanoma population. PMID: 28797232
- Data show that mutations in NRAS are associated with poor survival. PMID: 28446505
- s analyzed 421 samples from CLM patients for their all-RAS mutation status to compare the overall survival rate (OS), recurrence-free survival rate (RFS), and the pattern of recurrence between the patients with and without RAS mutations. PMID: 29194647
- The lack of KRAS, NRAS, BRAF, and PIK3CA mutation in our study may suggest that a subset of eyelid sebaceous carcinomas is unlike that of eyelid sebaceous carcinomas of western countries. PMID: 28551389
- in vitro assays confirmed that decreased miR-425 strongly induced inflammatory cytokines (interleukin [IL]-2 and interferon [IFN]-gamma) via N-Ras upregulation in the TCR signalling pathway in primary biliary cholangitis PMID: 28192189
- analysis of NRAS mutations in pulmonary Langerhans cell histiocytosis PMID: 27076591
- analysis of K-RAS and N-RAS mutations in testicular germ cell tumors PMID: 28426398
- KRAS mutations were rarely found together and those in codons 12 and 13 conferred poor prognosis. For BRAF, more c.1781A>G (p.D594G) colorectal cancers (CRC)carried RAS mutations [14% (3/21)] compared with c.1799T>A (p.V600E) CRCs.For NRAS, 5% (3/60) of codon 61 mutant colorectal cancers had KRAS mutations compared with 44% (10/23) of codons 12 and 13 mutant colorectal cancers PMID: 27815357
- Mutation analysis Iindicate NRAS as the most commonly mutated gene in myeloma patients followed by KRAS ( and BRAF. PMID: 27634910
- A rapid monophyletic evolution of melanoma subpopulations in response to targeted therapy that was not observed in non-targeted therapy was observed. NRAS mutations in BRAF mutated patient treated with a BRAF inhibitor were identified post-resistant samples. Sequence analysis showed that NRAS mutations co-occur with BRAF mutations in single cells, and are not mutually exclusive. PMID: 27791198
- studies suggest that ZDHHC9 may serve as a safe and effective target for developing therapies against NRAS-driven cancers PMID: 26493479
- MEK1 does not act as a general tumor suppressor in leukemogenesis. Rather, its effects strongly depend on the genetic context (RAS versus MYC-driven leukemia) and on the cell type involved. PMID: 27741509
- Report a synthetic lethal interaction of cetuximab in combination with MEK1/2 inhibition for the NRAS mutant subgroup of metastatic colorectal cancer. PMID: 27636997
- NRAS was identified as a direct target of let-7e and promoted oncogenesis in glioma stem cells. PMID: 27556696
- It may be that NRAS Q61R, found in around half of GCMNs,[9] is a less potent promoter of FGF23 than the HRAS mutations usually found in verrucous EN with CSHS, and requires a larger mutant clone to cause CSHS. PMID: 27900779
- Study showed that NRAS-mutation(+) colorectal cancer (CRC) had distinct epigenetic and clinicopathological features and significantly correlated with low-methylation epigenotypes. NRAS-mutation(+) CRC significantly correlated with less lymph vessel invasion, occurred preferentially in elder patients and at the distal colon, and showed relatively better prognosis, compared with KRAS-mutation(+) CRC. PMID: 28378457
- Oncogene NRAS G138R variant was identified as having a predicted damaging effect on protein function. PMID: 27121310
- NRASQ61R Mutation-specific Immunohistochemistry is Highly Specific for Either NRASQ61R or KRASQ61R Mutation in Colorectal Carcinoma. PMID: 26862952
- Results show that promoter mutations render telomerase reverse transcriptase (TERT) expression dependent on MAPK signal pathway activation due to oncogenic BRAF or NRAS mutations. PMID: 27449293
- Mutational activation of Kit-, Ras/Raf/Erk- and Akt- pathways indicate the biological importance of these pathways and their components as potential targets for therapy. PMID: 27391150
- An age-related increase on the frequency of NRAS mutations was observed. PMID: 27433783
- Melanomas from geographically different regions in New Zealand have markedly different mutation frequencies, in particular in the NRAS and EPHB6 genes, when compared to The Cancer Genome Atlas database or other populations. These data have implications for the causation and treatment of malignant melanoma in New Zealand. PMID: 27191502
- Data indicate acquired KRAS, NRAS or HRAS mutations in more than one third of patients after cetuximab exposure. PMID: 27119512
- Mutational status of NRAS, KRAS, and PTPN11 genes is associated with genetic/cytogenetic features in children with B-precursor acute lymphoblastic leukemia. PMID: 28853218
- The results demonstrated the lack of activity of anti-EGFRs in RAS(KRAS and NRAS) and BRAF wild-type, right-sided tumors, thus suggesting a potential role for primary tumor location in driving treatment choices PMID: 27382031
- Data indicate that BRAF, NRAS and C-KIT melanomas constitute distinct clinico-pathological entities. PMID: 29187493
- NRAS promotes interleukin-8-related chemokine secretion by tumor cells. PMID: 28341702
- MC1R genotype is associated with patient phenotypes with BRAF and NRAS mutations in melanoma PMID: 28842324
- Our data suggest that KRAS, NRAS, and BRAF mutations predict response to cetuximab treatment in metastatic colorectal cancer patients. PMID: 26989027
- Although recurrent NRAS mutations are present, the low mutation rate suggests that NRAS itself plays a minor role in the development of low-grade ovarian serous carcinoma. PMID: 28873354
- Results show that NRAS mRNA is a direct target of microRNA miR-340. PMID: 26799668
- complex signaling mechanisms that involve PREX2, PI3K/AKT/PTEN and downstream epigenetic machinery to deregulate expression of key cell cycle regulators PMID: 27111337
- he rapid tumor onset observed in this replication attempt, compared to the original study, makes the detection of accelerated tumor growth in PREX2 expressing NRAS(G12D) melanocytes extremely difficult. PMID: 28100394
- Mutations in KRAS, NRAS, and BRAF together occur in more than half of all colorectal cancer cases and are often associated with negative responses to the EGFR inhibitors cetuximab and panitumumab.guideline is clear that we should not be giving EGFR inhibitors to patients with RAS mutations and that patients with BRAF V600E mutations have a much worse prognosis PMID: 28249840
- KIT knockdown increased RAS/MAPK pathway activation in a BRAF(V600E)-mutant melanoma cell line. PMID: 28947418
- NRAS mutation is associated with response to therapy in rectal cancer. PMID: 28859058
- Age at diagnosis of follicular thyroid cancer (FTC) and frequency of extrathyroidal extension have changed over four decades; prevalence of RAS mutations has decreased; HRAS/NRAS (codon 61) and TERT (promoter) mutations may be associated with poor clinical outcomes in FTC, especially when two mutations coexist; this retrospective study was conducted in Seoul. PMID: 28864536
显示更多
收起更多
-
相关疾病:Leukemia, juvenile myelomonocytic (JMML); Noonan syndrome 6 (NS6); RAS-associated autoimmune leukoproliferative disorder (RALD); Melanocytic nevus syndrome, congenital (CMNS); Melanosis, neurocutaneous (NCMS); Keratinocytic non-epidermolytic nevus (KNEN); Thyroid cancer, non-medullary, 2 (NMTC2)
-
亚细胞定位:Cell membrane; Lipid-anchor; Cytoplasmic side. Golgi apparatus membrane; Lipid-anchor.
-
蛋白家族:Small GTPase superfamily, Ras family
-
数据库链接:
HGNC: 7989
OMIM: 137550
KEGG: hsa:4893
STRING: 9606.ENSP00000358548
UniGene: Hs.486502
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-