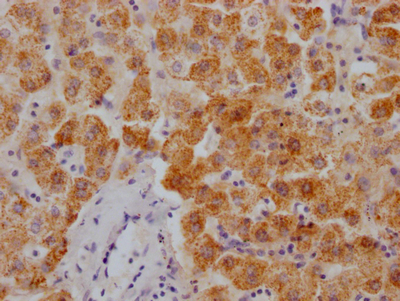
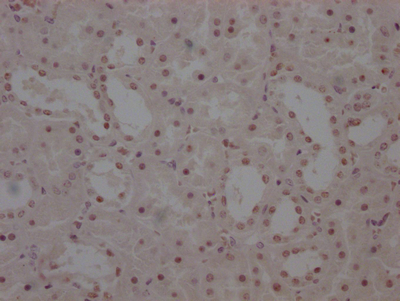
RPOD Antibody
-
货号:CSB-PA360419XA01ENV
-
规格:¥440
-
促销:
-
图片:
-
其他:
产品详情
-
Uniprot No.:P00579
-
基因名:RPOD
-
别名:RNA polymerase sigma factor RpoD (Sigma-70) rpoD alt b3067 JW3039
-
反应种属:Penaeus monodon
-
免疫原:Recombinant Escherichia coli (strain K12) RPOD protein
-
免疫原种属:Escherichia coli (strain K12)
-
标记方式:Non-conjugated
-
克隆类型:Polyclonal
-
抗体亚型:IgG
-
纯化方式:Protein G
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:Preservative: 0.03% Proclin 300
Constituents: 50% Glycerol, 0.01M PBS, pH 7.4 -
产品提供形式:Liquid
-
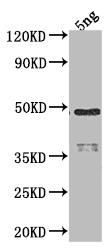
应用范围:ELISA, WB
-
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Sigma factors are initiation factors that promote the attachment of RNA polymerase to specific initiation sites and are then released. This sigma factor is the primary sigma factor during exponential growth. Preferentially transcribes genes associated with fast growth, such as ribosomal operons, other protein-synthesis related genes, rRNA- and tRNA-encoding genes and prfB.
-
基因功能参考文献:
- Counter-intuitively, intermolecular epistasis can alleviate the constraints of individual components, thereby increasing phenotypic variation that selection could act on and facilitating adaptive evolution. PMID: 29130883
- In contrast to NusG, which freely binds to RNA polymerase, RfaH exists in a structurally distinct autoinhibitory state in which the RNA polymerase-binding site is buried at the interface between two RfaH domains. PMID: 28605514
- Here the s establish that the primary sigma factor in Escherichia coli, sigma(70), can function as an elongation factor in vivo by loading directly onto the transcription elongation complex in trans. PMID: 26371553
- These data suggested that template strand single-stranded DNA competes with lipiarmycin for binding to RNA polymerase and that sigma(70) regions 1.2 and 3.2 attenuate lipiarmycin action by promoting DNA duplex opening. PMID: 26724534
- The sigma70-subunit of E. coli RNA polymerase (a small protein, being a part of RNA holoenzyme, and responsible for initiation of transcription of constitutive genes) is modeled at different ionic strengths PMID: 26841497
- Data suggest that RNAP sigma 70 interactions with DNA immediately downstream of the transcription bubble are particularly important for initiation of transcription. PMID: 25311862
- s conclude that bacteriophage interruption of mlrA and RpoS mutations provide major obstacles limiting curli expression and biofilm formation in most serotype O157 : H7 strains. PMID: 23744902
- where at least three regions of the 6S RNA must interact with Esigma(70) in a cooperative manner so as to ensure effective pRNA-dependent release. PMID: 24681966
- The study presents the initial steps of the 6S RNA - RNA polymerase complex decay. PMID: 23667906
- X-ray crystal structure of Escherichia coli RNA polymerase sigma70 holoenzyme PMID: 23389035
- Deliberately slowing the intrinsic 6S RNA release rate by nucleotide feeding experiments reveals that sigma70 ejection occurs abruptly once a pRNA length of 9 nucleotides is reached. PMID: 23118417
- study found that although Rob makes fewer interactions with sigma(70) R4 than SoxS, the 2 proteins make the same position-dependent interactions; Rob occludes sigma(70) R4 from binding the -35 hexamer, just as does SoxS; thus, the C-terminal domain does not substantially alter the way Rob interacts with sigma(70) R4 at class II promoters PMID: 22465792
- Data indicate that region 1.1 and, to a lesser extent, region 1.2 of sigma(70) subunit determine higher promoter complex stability of RNA polymerase (RNAP). PMID: 22605342
- changes in the structure of region 2 of the sigma(70) subunit have stronger effects on transcription pausing than on promoter recognition, likely by weakening the interactions of the sigma subunit with the core RNAP during transcription elongation. PMID: 22098235
- The strength of the -15 element in sigma(70) promoter inversely correlates with the strengths of the -35 element and -10 element. PMID: 21908667
- sigma70 drives yjbEFGH transcription and it also acts to repress its induction. PMID: 21396442
- Mutation of sigma(70) residue R451, which is highly conserved, results in reduced growth rate, consistent with a central role in promoter recognition. PMID: 21398630
- His180 substitutions influence several steps of transcription initiation, including core binding, promoter DNA recognition and open complex formation. PMID: 21451247
- Genetic evidence for a novel interaction between transcriptional activator soxs and region 4 of the sigma(70) subunit of RNA polymerase at class ii SoxS-dependent promoters in Escherichia coli PMID: 21195716
- SoxS is now the first example of an E. coli transcriptional activator that uses a single positive control surface to make specific protein-protein contacts with two different subunits of RNAP. PMID: 20595001
- Phenotypically, cells overproducing sigma(70) favours growth and reproduction at the expense of motility and damage protection. PMID: 19574956
- in vivo association of sigma(70) and NusA with elongating RNA polymerase is regulated by growth conditions PMID: 15596728
- analysis of promoter DNA melting by Thermus aquaticus and Escherichia coli RNA polymerases PMID: 15731103
- amino acid substitutions in region 4 of sigma(70) that have specific effects on the sigma(70) region 4/beta-flap RNA polymerase interaction, either weakening or strengthening it PMID: 15761057
- Establishment of downstream contacts leads to remodeling of upstream interactions between sigma70 and the -10 promoter element that might facilitate promoter escape and sigma release PMID: 15978618
- the recognition of the -35 region involves similar amino acid residues in regions 4.2 of E. coli sigma32 and sigma70 PMID: 16166539
- the core of promoter melting activity of RNA polymerase is localized to a very small subset of all promoter-polymerase contacts PMID: 16169843
- an important role of sigma region 3.2 in the binding of initiating substrates in RNA polymerase active center and in the process of promoter clearance PMID: 16690607
- Sigma region 1.2 acts as a core RNA polymerase-dependent allosteric switch that modulates non-template DNA strand recognition by sigma region 2 during transcription initiation and elongation. PMID: 17268549
- sigma(70) NCR/beta' interaction facilitates promoter escape and hinders early elongation pausing, in contrast to the sigma(2)/beta' coiled-coil interaction, which has opposite effects. PMID: 17332752
- experiments support a model in which the role of Rsd is primarily to sequester sigma(70), thereby increasing the levels of RNA polymerase containing the alternative sigma(38) factor PMID: 17351046
- The 2.6 A-resolution X-ray crystal structure of Rsd bound to sigma(70) domain 4 (sigma(70)(4)), the primary determinant for Rsd binding within sigma(70), was determined. PMID: 17681541
- the T429 of Sigma 70 acts as a competitor for the hydrogen bonding that stabilizes the highly conserved - 11A-T base pairs of the promoter DNA, thus facilitating initiation of strand separation at this particular position in the - 10 region. PMID: 18155246
- The tyrosine at residue 430 in region 2.3 of sigma(70) was shown to be involved in quenching the fluorescence of a 2-AP substituted at -11, presumably through a stacking interaction. PMID: 18976666
- Residues in sigma70 region 4.2, the C-terminus of the second helix, contribute significantly to 6S RNA binding but not to binding of a minimal promoter. PMID: 19538447
显示更多
收起更多
-
亚细胞定位:Cytoplasm.
-
蛋白家族:Sigma-70 factor family, RpoD/SigA subfamily
-
数据库链接:
KEGG: ecj:JW3039
STRING: 316385.ECDH10B_3242
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
-
-
-
-
-