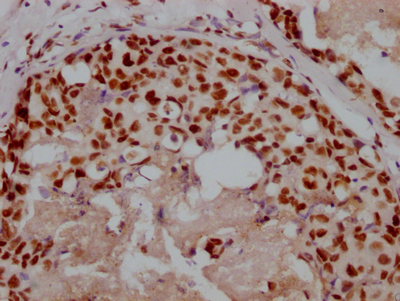
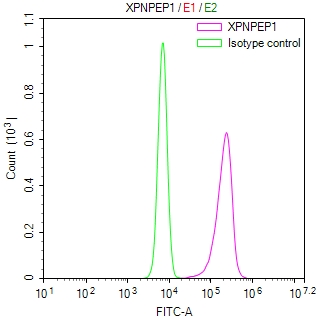
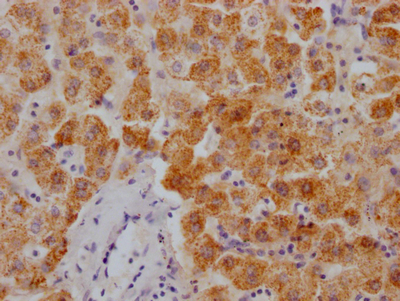
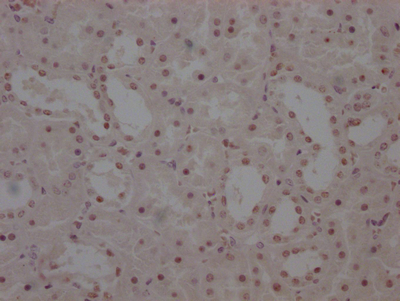
BRIP1 Recombinant Monoclonal Antibody
-
货号:CSB-RA268983A0HU
-
规格:¥1320
-
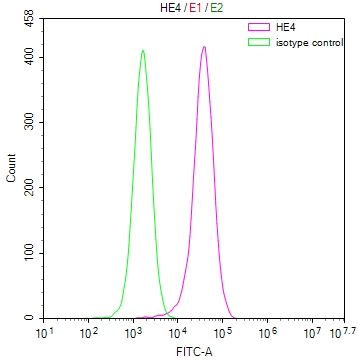
图片:
-
其他:
产品详情
-
Uniprot No.:Q9BX63
-
基因名:BRIP1
-
别名:Fanconi anemia group J protein (Protein FACJ) (EC 3.6.4.13) (ATP-dependent RNA helicase BRIP1) (BRCA1-associated C-terminal helicase 1) (BRCA1-interacting protein C-terminal helicase 1) (BRCA1-interacting protein 1), BRIP1, BACH1 FANCJ
-
反应种属:Human
-
免疫原:A synthesized peptide derived from Human BRIP1
-
免疫原种属:Homo sapiens (Human)
-
标记方式:Non-conjugated
-
克隆类型:Monoclonal
-
抗体亚型:Rabbit IgG
-
纯化方式:Affinity-chromatography
-
克隆号:27F4
-
浓度:It differs from different batches. Please contact us to confirm it.
-
保存缓冲液:Rabbit IgG in phosphate buffered saline, pH 7.4, 150mM NaCl, 0.02% sodium azide and 50% glycerol.
-
产品提供形式:Liquid
-
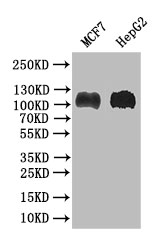
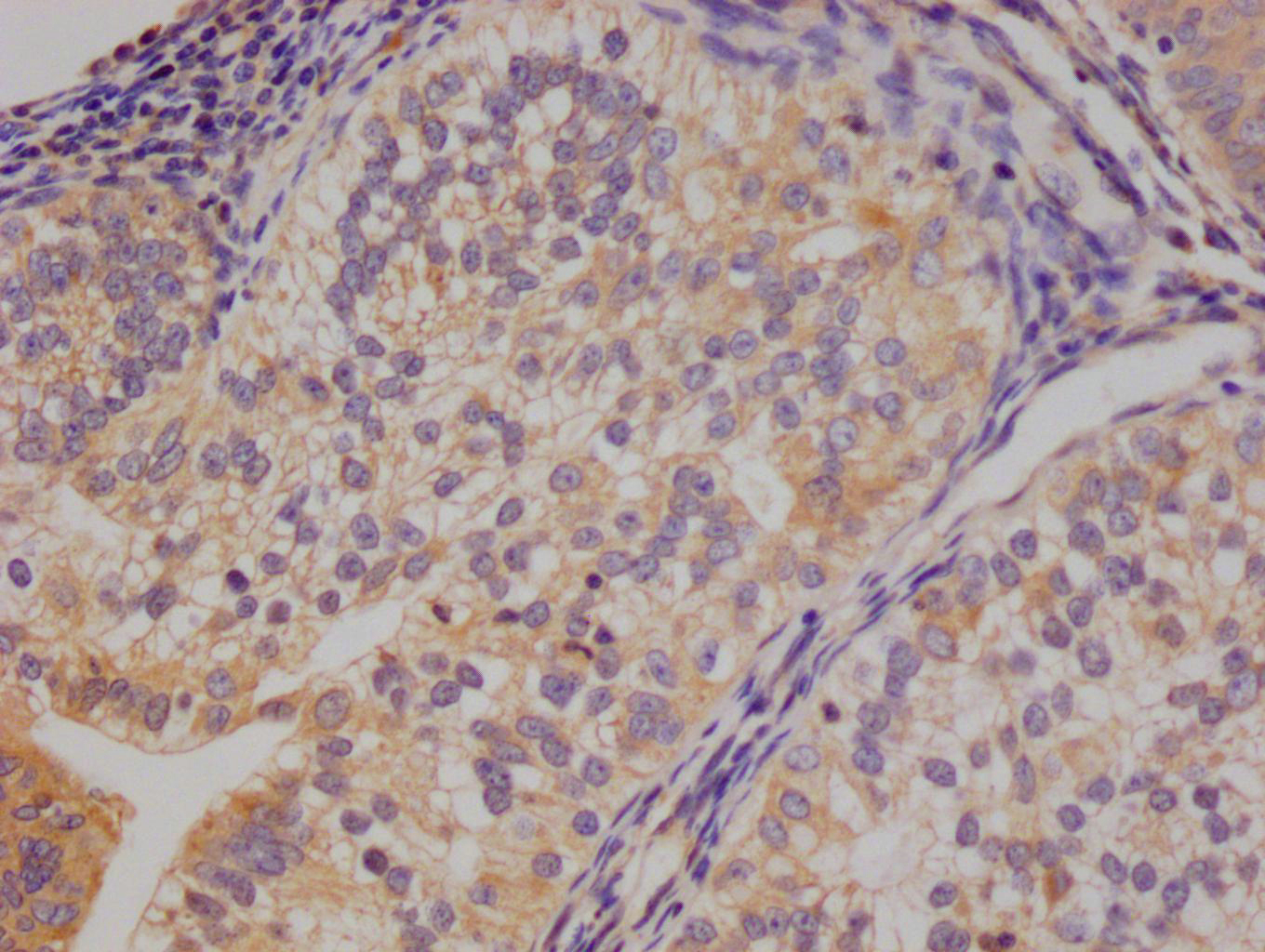
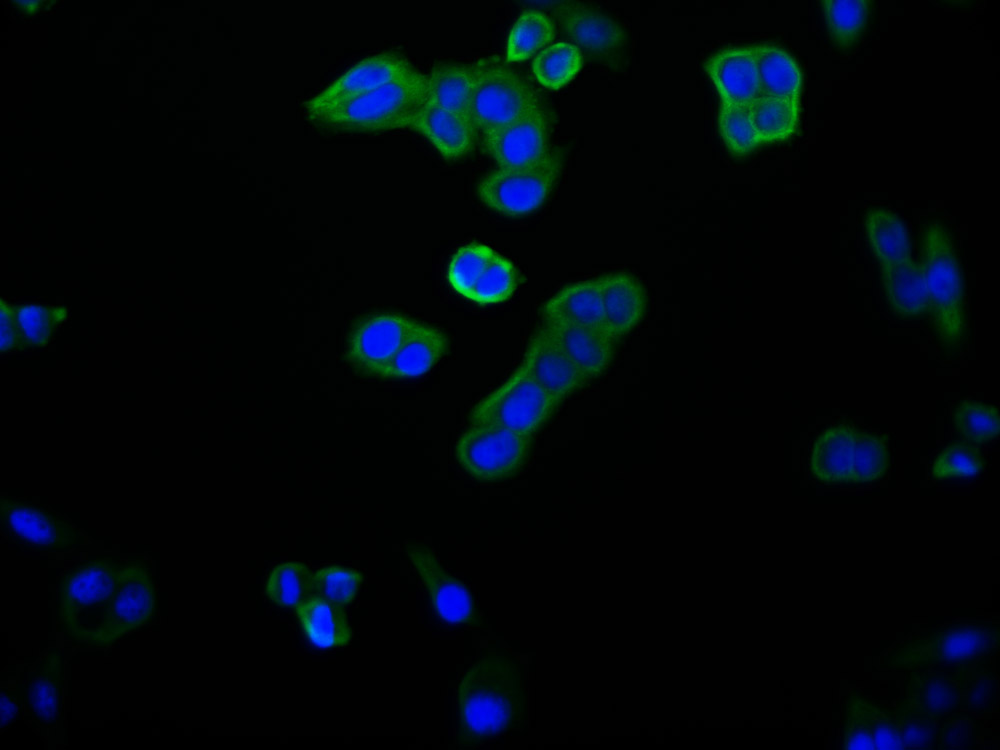
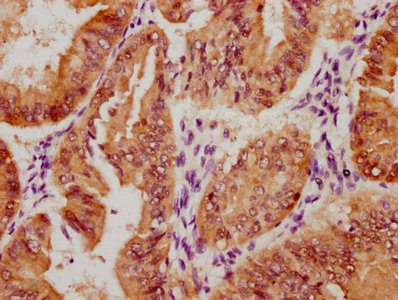
应用范围:ELISA, WB
-
推荐稀释比:
Application Recommended Dilution WB 1:500-1:2000 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:DNA-dependent ATPase and 5' to 3' DNA helicase required for the maintenance of chromosomal stability. Acts late in the Fanconi anemia pathway, after FANCD2 ubiquitination. Involved in the repair of DNA double-strand breaks by homologous recombination in a manner that depends on its association with BRCA1.
-
基因功能参考文献:
- Whole exome sequencing in triple negative breast cancer cases revealed BRIP1 rs552752779 (MAF: 75% vs. 6.25%, OR 45.00, 95% CI 9.43-243.32) are risk factors for triple negative breast cancer. PMID: 30136158
- Since BRCA1 and BACH1 mutations targeting the BRCA1-BACH1 interaction have been associated with breast cancer susceptibility, the results of the present study thus provide evidence for a novel role of BACH1 in tumour suppression. PMID: 22032289
- In a Chinese population, genetic variations in the BRIP1 gene are linked to increased risk for meningioma. PMID: 29581016
- LOH may predominantly indicate copy number gains in FANCF and losses in FANCG and BRIP1. Integration of copy number data and gene expression proved difficult as the available sample sets did not overlap. PMID: 28440438
- Because protein-truncating mutation in BRIP1 was not identified in the current study, it is unlikely that alterations in BRIP1 significantly contribute to the susceptibility of breast cancer in Korean patients. PMID: 26790966
- As an increasing number of clinically relevant FANCJ mutations are identified, understanding the mechanism whereby FANCJ mutation leads to diseases is critical. Mutational analysis of FANCJ will help us elucidate the pathogenesis and potentially lead to therapeutic strategies by targeting FANCJ. PMID: 27107905
- Cells expressing FANCJ pathological mutants exhibited defective sister chromatid recombination with an increased frequency of long-tract gene conversions. PMID: 28911102
- Ttruncating variants in BRIP1, and in particular p.Arg798Ter, are not associated with a substantial increase in breast cancer risk. PMID: 26921362
- Germline mutation in BRIP1 gene is associated with melanoma. PMID: 27074266
- diverse endogenous microsatellite signals were also lost upon replication stress after FANCJ depletion, and in FANCJ null patient cells. PMID: 27179029
- Findings collectively demonstrate that microRNA-543 exerts its oncogene function by directly targeting BRCA1-interacting protein 1 in cervical cancer. PMID: 28231728
- we showed the essential role of HP1 in regulating HR through BRCA1/BARD1-mediated accumulation of FANCJ and CtIP at DSB sites. This mechanism may affect tumorigenesis and chemosensitivity and is thus of high clinical significance. PMID: 27399284
- A variant at a potentially functional microRNA-binding site in BRIP1 was associated with risk of squamous cell carcinoma of the head and neck. PMID: 26711789
- BRIP1 might be the gene involved in the onset of breast cancer in families that do not show BRACA1/2 mutations (Review) PMID: 26709662
- Fancj helicase-deficient mice, while phenotypically resembling Fanconi anemia (FA), are also hypersensitive to replication inhibitors and predisposed to lymphoma PMID: 26637282
- FANCJ and BRCA2 share FANCD2's role in replication fork restart. PMID: 25659033
- Deleterious germline mutations in BRIP1 are associated with a moderate increase in the risk of EOC Epithelial Ovarian Cancer). PMID: 26315354
- our data point to the existence of a functional interplay between hMSH5 and FANCJ in double-strand break repair induced by replication stress. PMID: 26055704
- In coordination with BRCA1, FancJ promotes DNA damage-induced centrosome amplification in DNA damaged cells. PMID: 25483079
- Genetic variants in BRIP1 (BACH1) contribute to risk of nonsyndromic cleft lip with or without cleft palate. PMID: 25045080
- Our results suggest not only that FANCD2 regulates FANCJ chromatin localization but also that FANCJ is necessary for efficient loading of FANCD2 onto chromatin following DNA damage caused by mitomycin C treatment. PMID: 25070891
- The assessment of FANCD2, RAD51, BRCA1 and BRIP1 nuclear proteins could provide important information about the patients at risk for treatment failure. PMID: 24708616
- The interaction between TopBP1 and BACH1 is required for the extension of single-stranded DNA regions and RPA loading following replication stress, which is a prerequisite for the subsequent activation of replication checkpoint. PMID: 20159562
- we uncover an MLH1 clinical mutation with a leucine (L)-to-histidine (H) amino acid change at position 607 in hereditary nonpolyposis colon cancer that ablates MLH1 binding to FANCJ PMID: 20978114
- FANCJ localization by mismatch repair is vital to maintain genomic integrity after UV irradiation. PMID: 24351291
- Fanconi anemia group J (FANCJ) helicase partners with the single-stranded DNA-binding protein replication protein A (RPA) to displace BamHI-E111A bound to duplex DNA in a specific manner. PMID: 24895130
- FANCJ-MLH1 interaction is important for DNA damage responses. PMID: 24966277
- SNPs in BRIP1 are significantly associated with breast cancer. PMID: 24301948
- Loss of BRIP1 thus disrupts normal mammary morphogenesis and causes neoplastic-like changes, possibly via dysregulating multiple cellular signaling pathways functioning in the normal development of mammary glands. PMID: 24040146
- analysis of two Fanconi anemia patient mutations, R251C and Q255H, that are localized in helicase motif Ia of FANCJ PMID: 24573678
- The BRIP1 gene was screened for mutations in well-characterized, Finnish, high-risk hereditary breast and/or ovarian cancer individuals. PMID: 21356067
- The results strongly suggest that the decrease in FANCJ caused by 5-fluorouracil leads to an increase in sensitivity to oxaliplatin, indicating that the FANCJ protein plays an important role in the synergism of the combination of 5FU and oxaliplatin PMID: 22968820
- BRIP1 is a direct transcription target of FOXM1. Depletion of FOXM1 downregulates BRIP1 expression at the protein & mRNA levels. FOXM1 regulates BRIP1 expression to modulate epirubicin-induced DNA damage repair and drug resistance. PMID: 23108394
- BRIP1 gene polymorphisms play a role in cervical cancer in Chinese Han population. PMID: 23644138
- FANCJ helicase and MRE11 nuclease interact to facilitate the DNA damage response. PMID: 23530059
- variant alleles in two (Pro919Ser and G64A) of the three BRIP1 polymorphisms elicited no associations with breast cancer risk PMID: 23225146
- SNPs in the BRIP1 gene may influence cervical cancer susceptibility in a Chinese Han population. PMID: 23473757
- FANCJ expression may be a useful biomarker to predict sensitivity to 5-fluorouracil and prognosis in colorectal cancer. PMID: 22526901
- FANCJ phosphorylation is strongly induced by DNA-damaging agents. PMID: 23157317
- We show that acetylation at lysine 1249 is a critical regulator of FANCJ function during cellular DNA repair. PMID: 22792074
- the Q motif is essential for FANCJ enzymatic activity in vitro and DNA repair function in vivo PMID: 22582397
- Downregulation of BRIP1, a physiological partner of BRCA1 in the DNA repair pathway, triggers BRCA1 chromatin dissociation. PMID: 22137763
- six missense variants predicted to be causative were detected, one in BRIP1 and five in PALB2 PMID: 21409391
- BRIP1 gene variants may not play a relevant role in Male Breast Cancer predisposition PMID: 21165771
- FANCJ catalytic activity and its effect on BLM protein stability contribute to preservation of genomic stability and a normal response to replication stress. PMID: 21240188
- Molecular basis of BACH1/FANCJ recognition by TopBP1 in DNA replication checkpoint control. PMID: 21127055
- Genomic rearrangements of the BRIP1 gene is associated with breast cancer. PMID: 20567916
- FANCJ is recruited in response to replication stress and serves to link FANCD2 to BRCA1. PMID: 20676667
- recombinant FANCJ-A349P protein had reduced iron and was defective in coupling ATP hydrolysis and translocase activity to unwinding forked duplex or G-quadruplex DNA substrates or disrupting protein-DNA complexes PMID: 20639400
- FancB (FAAP95, FA core complex)showed differences in methylation in HNSCC. PMID: 20332657
收起更多
-
相关疾病:Breast cancer (BC); Fanconi anemia complementation group J (FANCJ)
-
亚细胞定位:Nucleus. Cytoplasm.
-
蛋白家族:DEAD box helicase family, DEAH subfamily
-
组织特异性:Ubiquitously expressed, with highest levels in testis.
-
数据库链接:
HGNC: 20473
OMIM: 114480
KEGG: hsa:83990
STRING: 9606.ENSP00000259008
UniGene: Hs.128903