Recombinant Escherichia coli Cell division topological specificity factor (minE)
-
中文名称:大肠杆菌minE重组蛋白
-
货号:CSB-YP358914ENV
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:大肠杆菌minE重组蛋白
-
货号:CSB-EP358914ENV
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:大肠杆菌minE重组蛋白
-
货号:CSB-EP358914ENV-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:大肠杆菌minE重组蛋白
-
货号:CSB-BP358914ENV
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:大肠杆菌minE重组蛋白
-
货号:CSB-MP358914ENV
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:minE
-
Uniprot No.:
-
别名:minE; b1174; JW1163Cell division topological specificity factor
-
种属:Escherichia coli (strain K12)
-
蛋白长度:full length protein
-
表达区域:1-88
-
氨基酸序列MALLDFFLSR KKNTANIAKE RLQIIVAERR RSDAEPHYLP QLRKDILEVI CKYVQIDPEM VTVQLEQKDG DISILELNVT LPEAEELK
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶点详情
-
功能:Prevents the cell division inhibition by proteins MinC and MinD at internal division sites while permitting inhibition at polar sites. This ensures cell division at the proper site by restricting the formation of a division septum at the midpoint of the long axis of the cell.
-
基因功能参考文献:
- This review summarizes the cell-free data supporting a molecular mechanism that explains the variety of patterns supported by MinD and MinE both in and out of the cell; with an emphasis on the recent appreciation for how multiple conformational states of MinE can drive oscillation by spatiotemporally regulating MinD interaction with the membrane. PMID: 29188788
- the dynamic association of the MTS with the beta-sheet is fine-tuned to balance MinE's need to sense MinD on the membrane with its need to diffuse in the cytoplasm, both of which are necessary for the oscillation PMID: 28652337
- Non-linear Min protein interactions generate harmonics that signal mid-cell division in Escherichia coli. PMID: 29040283
- membrane targeting sequence defines the lower limit of the concentration-dependent wavelength of Min protein patterns while restraining MinE's ability to stimulate MinD's ATPase activity. PMID: 28622374
- Two MinD mutant proteins, MinD(K11A) and MinD(DeltaMTS15), are unable to form polymers with MinC PMID: 25497011
- The MinCDE proteins prevent the formation of the Z-ring at poles by oscillating from pole to pole, thereby ensuring that the concentration of the Z-ring inhibitor, MinC, is lowest at the cell center. PMID: 24327341
- MinD and MinE interact with anionic phospholipids and regulate division plane formation in Escherichia coli PMID: 23012351
- The s find that N45 in MinD is essential for MinE-stimulated ATPase activity and suggest that it is a key residue affected by MinE binding. PMID: 22651575
- MinE shares common protein signatures with a group of membrane trafficking proteins in eukaryotic cells. These MinE signatures appear to affect membrane curvature. PMID: 21738659
- These results identify the MinD-dependent conformational changes in MinE that convert it from a latent to an active form and lead to a model of how MinE persists at the MinD-membrane surface. PMID: 21816275
- The binding sites for MinC and MinE overlap and reveals that they are at the MinD dimer interface. PMID: 21231967
- polymerization of MinE over MinD oligomers triggers dynamic instability leading to detachment from the membrane. PMID: 20212106
- report on stochastic switching of the MinD and MinE-protein distributions between the two cell halves in short Escherichia coli cells. PMID: 20308588
- MinE and the invariant Lys11 residue in the ATPase P-loop of MinD compete for binding to a common site within the MinD structure, thereby providing a plausible structural basis for the ability of MinE to activate the ATPase activity of MinD. PMID: 15458408
- Lysine 11 of MinD is required for interaction MinE; the three residues in helix 7 that interact with lysine 11 are not required for MinE binding but are required for MinE to stimulate the MinD ATPase PMID: 15629934
- simple five-reaction model of the Min system can explain all documented Min phenotypes, if stochastic kinetics and three-dimensional diffusion are accounted for PMID: 16846247
- In vivo measurements of the mobility of MinD and MinE using fluorescence correlation spectroscopy shows 2 distinct timescales; the diffusion constant of cytoplasmic MinE to be approximately 10 microm(2) s(-1) PMID: 17200601
- Min proteins undergo a periodic pole-to-pole oscillation that involves polymerization and ATPase activity of MinD under the PMID: 17483175
- MinCDE most often moved from one branch to another in an invariant order, following a nonreversing clockwise or counterclockwise direction over the time periods observed. PMID: 18178745
显示更多
收起更多
-
蛋白家族:MinE family
-
数据库链接:
KEGG: ecj:JW1163
STRING: 316385.ECDH10B_1227
Most popular with customers
-
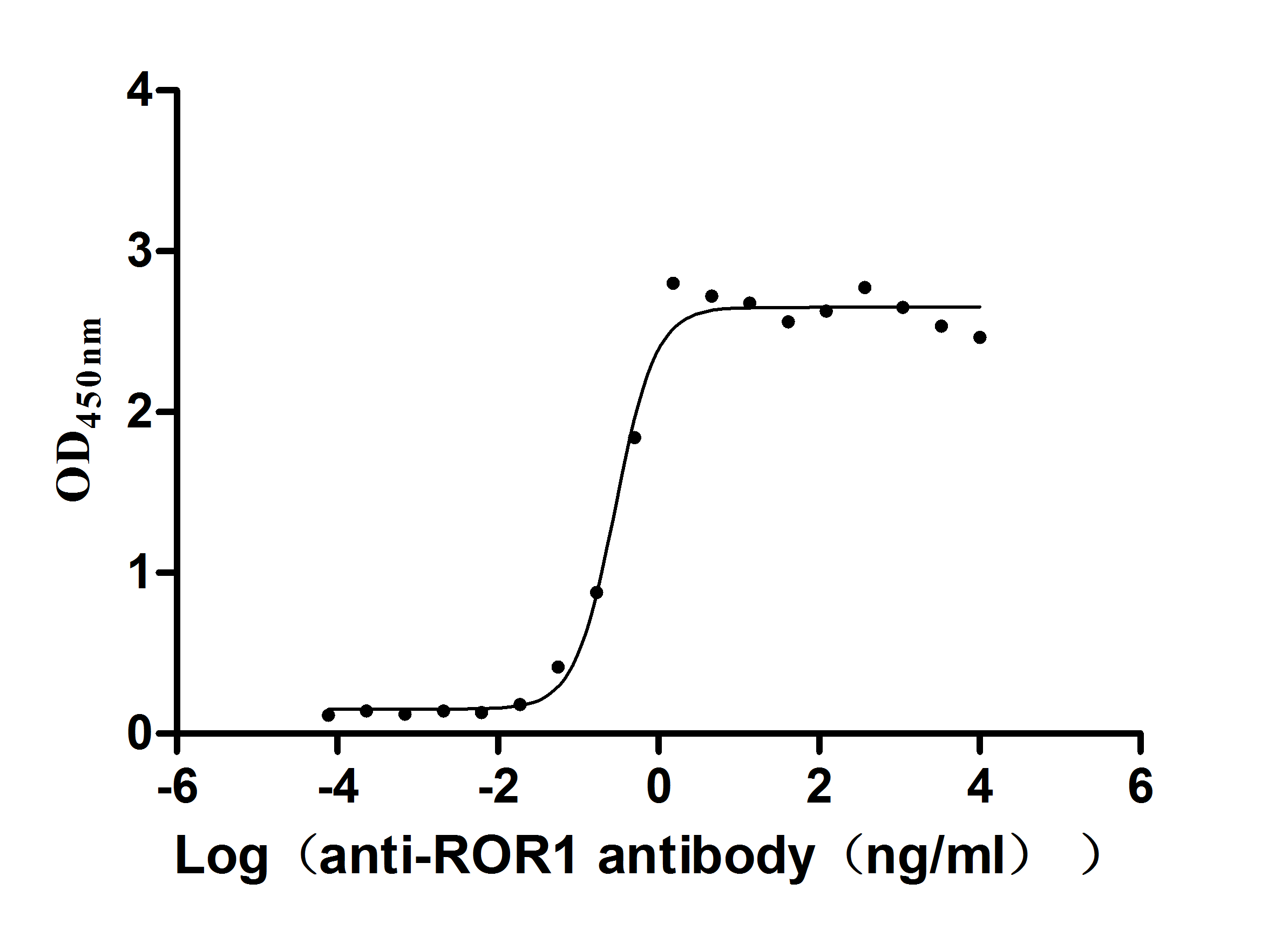
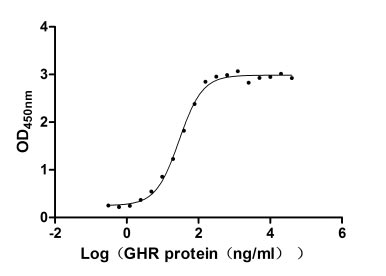
Recombinant Human Growth hormone receptor (GHR), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Glypican-3 (GPC3) (G537R), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
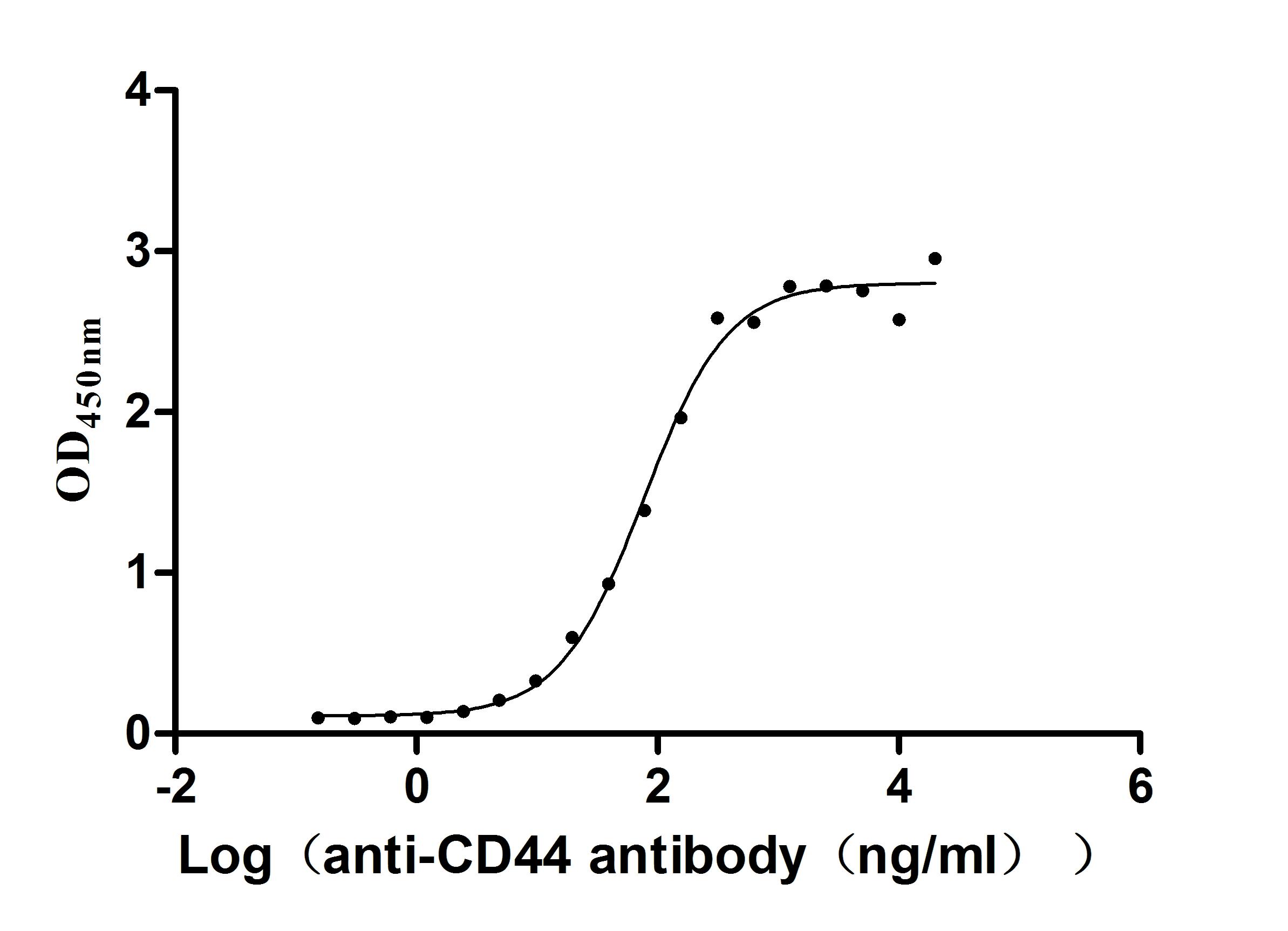
Recombinant Macaca fascicularis CD44 antigen (CD44), partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
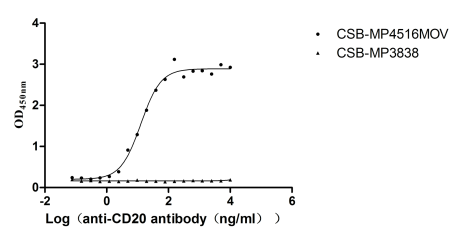
Recombinant Macaca fascicularis Membrane spanning 4-domains A1 (MS4A1)-VLPs (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
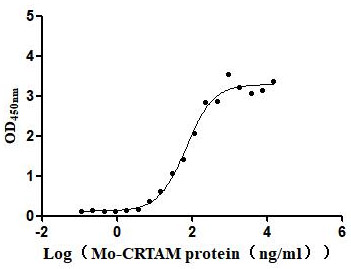
Recombinant Mouse Cell adhesion molecule 1 (Cadm1), partial (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
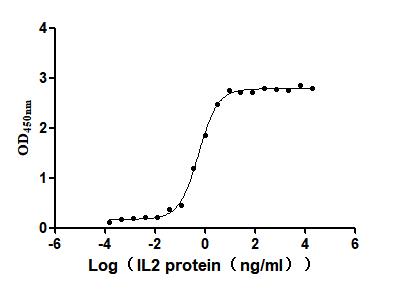
Recombinant Human Interleukin-2 (IL2) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-


-AC1.jpg)