Recombinant Human Eyes absent homolog 4 (EYA4)
-
货号:CSB-YP007909HU
-
规格:
-
来源:Yeast
-
其他:
-
货号:CSB-EP007909HU
-
规格:
-
来源:E.coli
-
其他:
-
货号:CSB-EP007909HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
货号:CSB-BP007909HU
-
规格:
-
来源:Baculovirus
-
其他:
-
货号:CSB-MP007909HU
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:
-
Uniprot No.:
-
别名:CMD1J; Deafness, autosomal dominant 10; DFNA 10; DFNA10; dJ78N10.1 (eyes absent (Drosophila) homolog 4); dJ78N10.1 (eyes absent); EYA 4; eya4; EYA4_HUMAN; Eyes absent 4; Eyes absent homolog 4 (Drosophila) ; Eyes absent homolog 4; HGNC:3522; OTTHUMP00000040267
-
种属:Homo sapiens (Human)
-
蛋白长度:Full length protein
-
表达区域:1-639
-
氨基酸序列MEDSQDLNEQ SVKKTCTESD VSQSQNSRSM EMQDLASPHT LVGGGDTPGS SKLEKSNLSS TSVTTNGTGG ENMTVLNTAD WLLSCNTPSS ATMSLLAVKT EPLNSSETTA TTGDGALDTF TGSVITSSGY SPRSAHQYSP QLYPSKPYPH ILSTPAAQTM SAYAGQTQYS GMQQPAVYTA YSQTGQPYSL PTYDLGVMLP AIKTESGLSQ TQSPLQSGCL SYSPGFSTPQ PGQTPYSYQM PGSSFAPSST IYANNSVSNS TNFSGSQQDY PSYTAFGQNQ YAQYYSASTY GAYMTSNNTA DGTPSSTSTY QLQESLPGLT NQPGEFDTMQ SPSTPIKDLD ERTCRSSGSK SRGRGRKNNP SPPPDSDLER VFVWDLDETI IVFHSLLTGS YAQKYGKDPP MAVTLGLRME EMIFNLADTH LFFNDLEECD QVHIDDVSSD DNGQDLSTYS FATDGFHAAA SSANLCLPTG VRGGVDWMRK LAFRYRRVKE LYNTYKNNVG GLLGPAKRDA WLQLRAEIEG LTDSWLTNAL KSLSIISTRS NCINVLVTTT QLIPALAKVL LYSLGGAFPI ENIYSATKIG KESCFERIMQ RFGRKVVYVV IGDGVEEEQA AKKHNMPFWR ISSHSDLLAL HQALELEYL
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:Tyrosine phosphatase that specifically dephosphorylates 'Tyr-142' of histone H2AX (H2AXY142ph). 'Tyr-142' phosphorylation of histone H2AX plays a central role in DNA repair and acts as a mark that distinguishes between apoptotic and repair responses to genotoxic stress. Promotes efficient DNA repair by dephosphorylating H2AX, promoting the recruitment of DNA repair complexes containing MDC1. Its function as histone phosphatase probably explains its role in transcription regulation during organogenesis. May be involved in development of the eye.
-
基因功能参考文献:
- Overexpression of EYA4 enhanced glioma cell proliferation, and EYA4 suppressed the expression of p27Kip1 directly in these cells. PMID: 30231237
- Although the clinical patient outcome of our 38 Colorectal Cancer patients was not associated with EYA4 promoter hypermethylation, the high frequency of this methylation and its high sensitivity and specificity to neoplastic cells may qualify EYA4 promoter methylation as a potential candidate screening marker in Iranian population and may help to improve early detection of CRC. PMID: 29436791
- Eyes absent homolog 4 (Drosophila) protein (EYA4) is frequently hypermethylated in esophageal squamous cell carcinoma (ESCC) and may function as a tumor suppressor gene in the development of ESCC. PMID: 29660222
- identified novel EYA4 mutation can be considered responsible of the hearing loss observed in the proband and her father, while a dual molecular diagnosis was reached in the relatives co-segregating the EYA4 and the PAX3 mutations PMID: 29287889
- EYA4 hypermethylation is associated with colorectal cancer. PMID: 28351398
- EYA4 functions as tumor suppressor gene in pancreatic ductal adenocarcinoma via repressing beta-catenin/ID2 activation, and was an independent prognostic factor in PDAC. PMID: 27378242
- Low expression of EYA4 is associated with oral cancer. PMID: 27015871
- We discovered two genome-wide significant SNPs. The first was novel and near ISG20. The second was in TRIOBP, a gene previously associated with prelingual nonsyndromic hearing loss. Motivated by our TRIOBP results, we also looked at exons in known hearing loss genes, and identified two additional SNPs, rs2877561 in ILDR1 and rs9493672 in EYA4 (at a significance threshold adjusted for number of SNPs in those regions). PMID: 27764096
- Locus polymorphism of rs3813346 was associated with the risk of developing noise-induced hearing loss in the dominance model, the codominance model and the addictive model. Generalized multiple dimensionality reduction indicated that the combined interaction of the 2 loci-rs3813346 and rs9493627-significantly affected the incidence of noise-induced hearing loss. PMID: 27613755
- Up to now, merely 7 loci have been linked to mid-frequency hearing loss. Only four genetic mid-frequency deafness genes, namely, DFNA10 (EYA4), DFNA8/12 (TECTA), DFNA13 (COL11A2), DFNA44 (CCDC50), have been reported to date. [review] PMID: 27142990
- study identified EYA4 gene as targets for AML1-ETO and indicated it as a novel tumor suppressor gene. In addition, we provided evidence that EYA4 gene might be a novel therapeutic target and a potential candidate for treating AML1-ETO+ t (8;21) AML. PMID: 27231175
- Loss of EYA4 expression is associated with intrahepatic cholangiocarcinoma. PMID: 27469137
- The identification of a novel EYA4 truncation mutation associated with DFNA10, rather than syndromic hearing loss, supports a previously reported genotype-phenotype correlation in this gene. PMID: 26015337
- results implicate Eya4/Six1 regulates normal cardiac function via p27/casein kinase-2alpha/histone deacetylase 2 and indicate that mutations within this transcriptional complex and signaling cascade lead to the development of cardiomyopathy. PMID: 26499333
- In a Dutch family with c.464del EYA4 mutation, hearing impairment begins as a mid-frequency hearing impairment in childhood and develops into a high-frequency, moderate hearing impairment later in life. PMID: 26331839
- Genetic variations in the EYA4, GRHL2 and DFNA5 genes and their interactions with occupational noise exposure may play an important role in the incidence of noise-induced hearing loss (NIHL). PMID: 26400775
- analysis of an EYA4 mutation causing hearing loss in a Chinese DFNA family PMID: 25963406
- A novel missense mutation c.1643C>G (p.T548R) in EYA4 may cause autosomal dominant non-syndromic hearing impairment. PMID: 25809937
- EYA4 mutations are associated with autosomal dominant non-syndromic hearing loss. PMID: 25781927
- Exome Sequencing Identifies a Mutation in EYA4 as a Novel Cause of Autosomal Dominant Non-Syndromic Hearing Loss PMID: 25961296
- EYA4 methylation may be identified in stool samples. PMID: 25620232
- Results provide molecular and clinical information in order to gain improved understanding of the pathogenesis of DFNA10 protein EYA4 mutations and the genotypephenotype correlations of DFNA10 hearing loss. PMID: 25242383
- Autosomal dominant hearing impairment due to a novel EYA4 frameshift mutation:a novel heterozygous frame-shift mutation (c.579_580insTACC, p.(Asp194Tyrfs*52)) in EYA4 was identified that truncates the so-called variable region of the protein. PMID: 25681523
- Low EYA4 expression is associated with hepatocellular carcinoma. PMID: 24306662
- Work shows a clear role for EYA4 as a putative tumor suppressor genes in non-small-cell lung cancer. PMID: 24096489
- High methylation of the EYA4 gene is associated with ulcerative colitis with colorectal cancer. PMID: 23867875
- Serum methylation levels of TAC1, SEPT9, and EYA4 were significant discriminants between stage I colorectal cancer and healthy controls. PMID: 23862763
- Observational study and genome-wide association study of gene-disease association, gene-environment interaction, and pharmacogenomic / toxicogenomic. (HuGE Navigator) PMID: 21061259
- Clinical trial of gene-disease association and gene-environment interaction. (HuGE Navigator) PMID: 20379614
- EYA4 and hTERT mRNA expression increased with the severity of esophageal pathological changes and may be useful for identifying high-risk endoscopy candidates or for monitoring changes in premalignant esophageal lesions. PMID: 19939248
- Findings identify a role of EYA4 and possibly interacting SIX and DACH proteins in MPNSTs and suggest the EYA4 pathway as a rational therapeutic target. PMID: 19901965
- Mutation analysis of the EYA4 gene, which maps to 6q22.3, revealed an insertion of 4 bp (1558insTTTG) in affected family members with hereditary hearing impairment PMID: 12477971
- Mutation in the transcriptional coactivator EYA4 causes dilated cardiomyopathy and sensorineural hearing loss PMID: 15735644
- Results show the first definitive cardiac evaluations of DFNA10 hearing loss to support a correlation of EYA4 mutation with/without the of dilated cardiomyopathy, and will facilitate the counseling of patients with these phenotypes and EYA4 mutations. PMID: 17567890
- Study is the first report of a point mutation in EYA4 that is hypothesized to lead to aberrant pre-mRNA splicing and human disease. PMID: 17568404
- Mice lacking the orthologous gene have severe hearing deficits, suggesting that some human otitis media susceptibility reflects underlying genetic predisposition in genes such as this one. PMID: 18219393
显示更多
收起更多
-
相关疾病:Deafness, autosomal dominant, 10 (DFNA10); Cardiomyopathy, dilated 1J (CMD1J)
-
亚细胞定位:Cytoplasm. Nucleus.
-
蛋白家族:HAD-like hydrolase superfamily, EYA family
-
组织特异性:Highly expressed in heart and skeletal muscle.
-
数据库链接:
HGNC: 3522
OMIM: 601316
KEGG: hsa:2070
STRING: 9606.ENSP00000347294
UniGene: Hs.596680
Most popular with customers
-
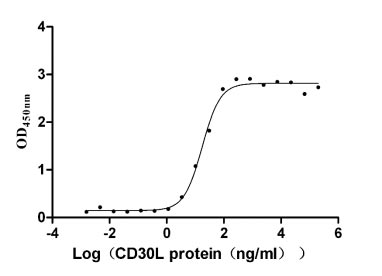
Recombinant Human Tumor necrosis factor receptor superfamily member 8 (TNFRSF8), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
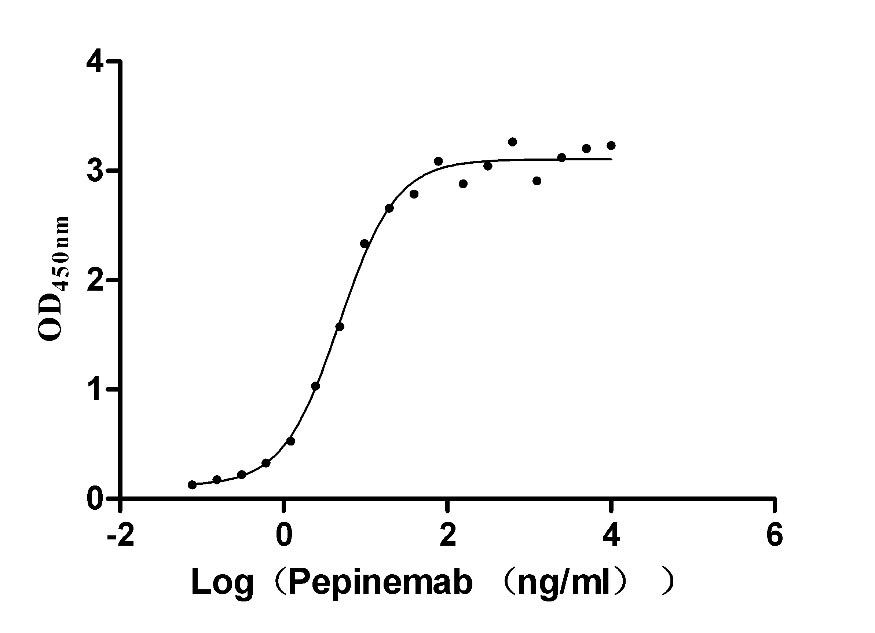
Recombinant Human Semaphorin-4D (SEMA4D), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
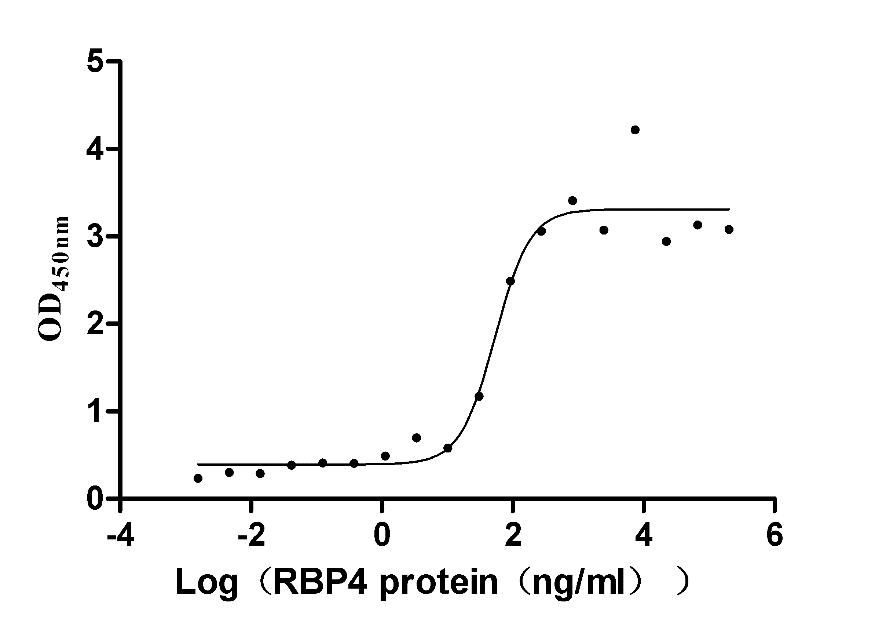
Recombinant Mouse Retinol-binding protein 4 (Rbp4) (Active)
Express system: Mammalian cell
Species: Mus musculus (Mouse)
-
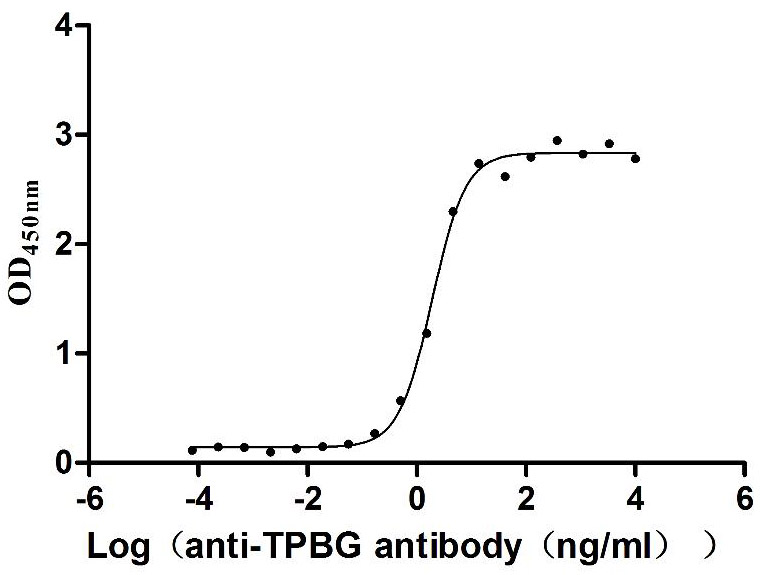
Recombinant Macaca fascicularis Trophoblast glycoprotein (TPBG), partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
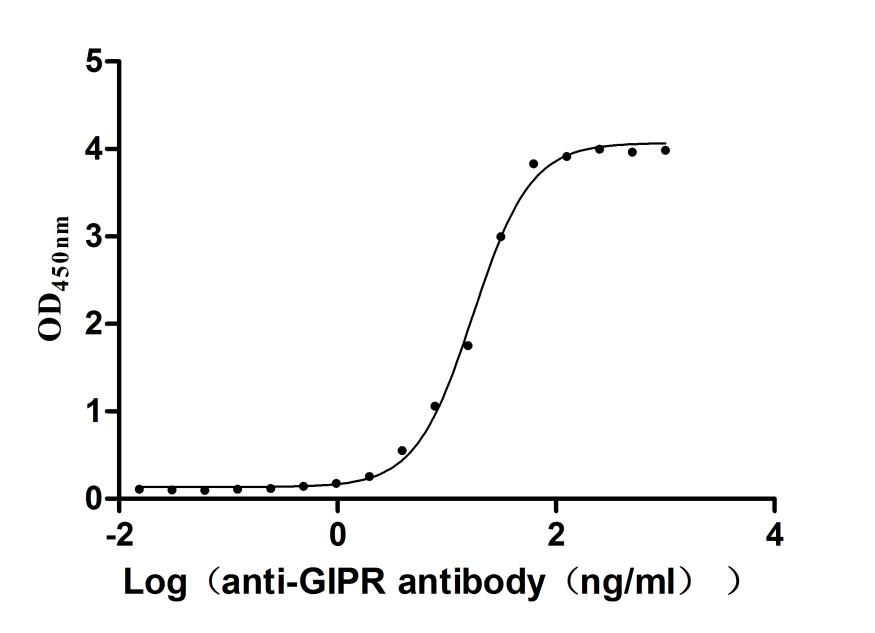
Recombinant Human Gastric inhibitory polypeptide receptor(GIPR),partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human C-C chemokine receptor type 9 (CCR9)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
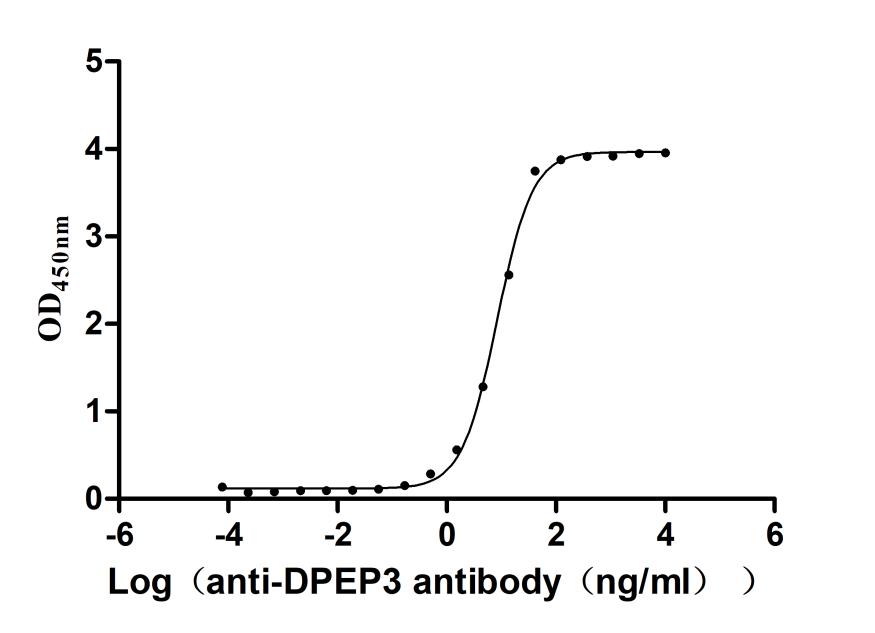
Recombinant Macaca fascicularis Dipeptidase 3(DPEP3) (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)