Recombinant Human Histone H3.3 (H3F3A), partial
-
货号:CSB-EP010109HUe0
-
规格:¥1344
-
图片:
-
其他:
产品详情
-
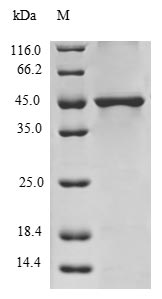
纯度:Greater than 90% as determined by SDS-PAGE.
-
基因名:H3F3A
-
Uniprot No.:
-
别名:H3 histone family 3A; H3 histone family 3B; H3 histone; family 3B (H3.3B); H3.3; H3.3A; H3.3B; H33_HUMAN; H3F3; H3F3A; H3f3b; Histone H3.3; Histone H3.3Q; Histone H3.A; Histone H3.B; MGC87782; MGC87783
-
种属:Homo sapiens (Human)
-
蛋白长度:partial
-
来源:E.coli
-
分子量:42.2kDa
-
表达区域:2-136aa
-
氨基酸序列ARTKQTARKSTGGKAPRKQLATKAARKSAPSTGGVKKPHRYRPGTVALREIRRYQKSTELLIRKLPFQRLVREIAQDFKTDLRFQSAAIGALQEASEAYLVGLFEDTNLCAIHAKRVTIMPKDIQLARRIRGERA
Note: The complete sequence including tag sequence, target protein sequence and linker sequence could be provided upon request. -
蛋白标签:N-terminal GST-tagged
-
产品提供形式:Liquid or Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
缓冲液:Tris-based buffer,50% glycerol
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet & COA:Please contact us to get it.
相关产品
靶点详情
-
功能:Variant histone H3 which replaces conventional H3 in a wide range of nucleosomes in active genes. Constitutes the predominant form of histone H3 in non-dividing cells and is incorporated into chromatin independently of DNA synthesis. Deposited at sites of nucleosomal displacement throughout transcribed genes, suggesting that it represents an epigenetic imprint of transcriptionally active chromatin. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
-
基因功能参考文献:
- We conclude that the Clival GCT is genetically defined by somatic mutation in the H3F3A gene, linking it to the GCT of long bones. PMID: 29609578
- overexpression of H3F3A, encoding H3.3, is associated with lung cancer progression and promotes lung cancer cell migration by activating metastasis-related genes PMID: 27694942
- we describe the presence of the mutation p.K27M of H3F3A (H3.3K27M) in two tumours of young patients with classical histopathology of ganglioglioma PMID: 27219822
- H3F3 mutations are sensitive and specific markers of giant cell tumors of the bone and chondroblastomas PMID: 28059095
- H3F3A is the most frequently mutated giant cell tumor of bone driver gene. H3F3A mutations are not present in atypical giant cell tumor of bone. PMID: 28545165
- H3F3A mutational testing may be a useful adjunct to differentiate giant cell tumor of bone from giant cell-rich sarcoma. PMID: 28899740
- Report H3F3A/B mutations in cell tumors of bone, chondroblastomas, and aneurysmal bone cysts. PMID: 28882701
- Data suggest that H3K9ac (histone 3 lysine 9 acetylation) serves as a substrate for direct binding of the SEC (super elongation complex) to chromatin; at select gene promoters, H3K9ac loss or SEC depletion appears to reduce gene expression. PMID: 28717009
- We determined the incidence of H3.3 G34 mutations in primary malignant bone tumors as assessed by genotype and H3.3 G34W immunostaining PMID: 28505000
- The kinase activity of Aurora B on serine 31 of histone H3.3 was biochemically confirmed with nucleosomal substrates in vitro. PMID: 28137420
- This study showed that heterozygous K27M mutations in H3F3A (n = 4) or HIST1H3B (n = 3) across all primary, contiguous, and metastatic tumor sites in all Diffuse intrinsic pontine glioma. PMID: 26727948
- Study examined the relationship of K27M mutations in the distinct histone H3 variants (i.e. HIST1H3B and H3F3A) with specific pontine glioma biology PMID: 26399631
- study found spinal high-grade gliomas in children and adults frequently harbor H3F3A (K27M) mutations PMID: 26231952
- H3F3A and H3F3B mutation analysis appears to be a highly specific, although less sensitive, diagnostic tool for the distinction of GCTB and chondroblastoma from other giant cell-containing tumors. PMID: 26457357
- we describe three interesting cases of paediatric glial and glioneuronal tumours harbouring both BRAF V600E and H3F3A K27M mutations. PMID: 25389051
- our observations further extend the knowledge of H3F3A mutation and its location in pediatric glioblastomas PMID: 25479829
- The CENP-A/histone H3.3 nucleosome forms an unexpectedly stable structure and allows the binding of the essential centromeric protein, CENP-C, which is ectopically mislocalized in the chromosomes of CENP-A overexpressing tumor cells. PMID: 25408271
- On the basis of our findings, H3F3A p.Gly34 Trp or p.Gly34 Leu mutations are not a frequent event in CGCL. PMID: 25442495
- These results suggest that immunohistochemical detection of H3.3 K27M is a sensitive and specific surrogate for the H3F3A K27M mutation and defines a prognostically poor subset of pediatric glioblastomas. PMID: 25200322
- this study identifies an H3.3K36me3-specific reader and a regulator of intron retention and reveals that BS69 connects histone H3.3K36me3 to regulated RNA splicing, providing significant, important insights into chromatin regulation of pre-mRNA processing. PMID: 25263594
- The mutually exclusive associations of HDAC1/p300, p300/histone, and HDAC1/histone on chromatin contribute to the dynamic regulation of histone acetylation. PMID: 24722339
- Loss of H3.3 from pericentromeric heterochromatin upon DAXX or PML depletion suggests that the targeting of H3.3 to PML-NBs is implicated in pericentromeric heterochromatin organization. PMID: 24200965
- These data suggest that adult brainstem gliomas differ from adult supratentorial gliomas. In particular, histone genes HIST1H3B (K27M) ) mutations are frequent in adult brainstem gliomas. PMID: 24242757
- H3F3A K27M mutation is associated with thalamic gliomas. PMID: 24285547
- The results of this study indicate that H3F3A K27M mutant GBMs show decreased H3K27me3 that may be of both diagnostic and biological relevance. PMID: 23414300
- H3F3A exon 2 mutation analyzed in solid tumors from 1351 South Korean patients PMID: 23758177
- A remarkable picture of tumor type specificity for histone H3.3 driver alterations emerges, indicating that histone H3.3 residues, mutations and genes have distinct functions. PMID: 24162739
- Reduced H3K27me3 and/or DNA hypomethylation are the major driving forces of activated gene expression in K27M mutant pediatric high-grade gliomas. PMID: 24183680
- All reported H3.3 mutations identified in human tumors have been in the H3F3A gene leading to single codon changes within the N-terminal tail of the H3.3 protein. [Review] PMID: 24229707
- This study suggested that none of H3.3 G34R mutated tumors presented primitive neuroectodermal tumors of central nervous system and pediatric glioblastomas. PMID: 23354654
- diffuse intrinsic pontine gliomas containing K27M mutation display lower overall amounts of H3 with trimethylated lysine 27(H3K27me3);H3K27M inhibits enzymatic activity of Polycomb repressive complex 2 through interaction with the EZH2 subunit; propose a model where aberrant epigenetic silencing through H3K27M-mediated inhibition of PRC2 activity promotes gliomagenesis PMID: 23539183
- Low frequency of H3.3 mutations in myelodysplastic syndromes patients. PMID: 23660862
- indicate that H3.3K27M mutation reprograms epigenetic landscape and gene expression, which may drive tumorigenesis PMID: 23603901
- H3F3A K27M mutations occur exclusively in pediatric diffuse high-grade astrocytomas PMID: 23429371
- Somatic mutation of H3F3A, a chromatin remodeling gene, is rare in acute leukemias and non-Hodgkin lymphoma. PMID: 23116151
- K27M mutation in H3.3 is universally associated with short survival in diffuse intrinsic pontine gliomas, while patients wild-type for H3.3 show improved survival. PMID: 22661320
- demonstrate that the two H3F3A mutations give rise to glioblastomas in separate anatomic compartments, with differential regulation of transcription factors OLIG1, OLIG2, and FOXG1, possibly reflecting different cellular origins PMID: 23079654
- recurrent mutations in a regulatory histone in humans; data suggest that defects of the chromatin architecture underlie paediatric and young adult GBM pathogenesis PMID: 22286061
- discussion of the importance of H3.3 deposition as a salvage pathway to maintain chromatin integrity PMID: 22195966
- Part of multiple H3.3-specific histone chaperone complexes PMID: 21047901
- Studies indicate that H3.3 accomplishes a surprising variety of cellular and developmental processes. PMID: 20153629
- Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex PMID: 11850414
- analysis of histone posttranslational modifications on H3.1 and H3.3 PMID: 17052464
- Data reveal that TPA activates transcription of TBX2 through activating MSK1, which leads to an increase in phosphorylated histone H3 and the recruitment of Sp1 to the TBX2 gene. PMID: 19633291
显示更多
收起更多
-
相关疾病:Glioma (GLM)
-
亚细胞定位:Nucleus. Chromosome.
-
蛋白家族:Histone H3 family
-
数据库链接:
HGNC: 4764
OMIM: 137800
KEGG: hsa:3020
STRING: 9606.ENSP00000355778
UniGene: Hs.180877
Most popular with customers
-
Recombinant Human Tumor necrosis factor receptor superfamily member 14 (TNFRSF14), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human CD40 ligand (CD40LG), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Tumor necrosis factor receptor superfamily member 5 (CD40), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Leukemia inhibitory factor (LIF) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Intestinal-type alkaline phosphatase (ALPI) (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Claudin-3 (CLDN3)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Macaca fascicularis zymogen granule protein 16 homolog B (ZG16B) (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
Recombinant Human Desmoglein-2 (DSG2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)