Recombinant Human Histone-lysine N-methyltransferase SETDB1 (SETDB1)
-
货号:CSB-YP619960HU
-
规格:
-
来源:Yeast
-
其他:
-
货号:CSB-EP619960HU-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
货号:CSB-BP619960HU
-
规格:
-
来源:Baculovirus
-
其他:
-
货号:CSB-MP619960HU
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:
-
Uniprot No.:
-
别名:AU022152; EC 2.1.1.43; ERG-associated protein with SET domain; ESET; H3 K9 HMTase 4; H3-K9-HMTase 4; H3-K9-HMTase4; Histone H3 K9 methyltransferase 4; Histone H3-K9 methyltransferase 4; Histone-lysine N-methyltransferase SETDB1; KG1T; KIAA0067; KMT1E; Lysine N-methyltransferase 1E; MGC90670; mKIAA0067; SET domain bifurcated 1; Set domain protein; bifurcated; 1; SETB1_HUMAN; Setdb1
-
种属:Homo sapiens (Human)
-
蛋白长度:full length of Isoform 2
-
表达区域:1-397
-
氨基酸序列MSSLPGCIGLDAATATVESEEIAELQQAVVEELGISMEELRHFIDEELEKMDCVQQRKKQLAELETWVIQKESEVAHVDQLFDDASRAVTNCESLVKDFYSKLGLQYRDSSSEDESSRPTEIIEIPDEDDDVLSIDSGDAGSRTPKDQKLREAMAALRKSAQDVQKFMDAVNKKSSSQDLHKGTLSQMSGELSKDGDLIVSMRILGKKRTKTWHKGTLIAIQTVGPGKKYKVKFDNKGKSLLSGNHIAYDYHPPADKLYVGSRVVAKYKDGNQVWLYAGIVAETPNVKNKLRFLIFFDDGYASYVTQSELYPICRPLKKTWEDIEDISCRDFIEEYVTAYPNRPMVLLKSGQLIKTEWEGTWWKSRVEEVDGSLVRILFLVLFFSTILEAEVGGGGT
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
相关产品
靶点详情
-
功能:Histone methyltransferase that specifically trimethylates 'Lys-9' of histone H3. H3 'Lys-9' trimethylation represents a specific tag for epigenetic transcriptional repression by recruiting HP1 (CBX1, CBX3 and/or CBX5) proteins to methylated histones. Mainly functions in euchromatin regions, thereby playing a central role in the silencing of euchromatic genes. H3 'Lys-9' trimethylation is coordinated with DNA methylation. Required for HUSH-mediated heterochromatin formation and gene silencing. Forms a complex with MBD1 and ATF7IP that represses transcription and couples DNA methylation and histone 'Lys-9' trimethylation. Its activity is dependent on MBD1 and is heritably maintained through DNA replication by being recruited by CAF-1. SETDB1 is targeted to histone H3 by TRIM28/TIF1B, a factor recruited by KRAB zinc-finger proteins. Probably forms a corepressor complex required for activated KRAS-mediated promoter hypermethylation and transcriptional silencing of tumor suppressor genes (TSGs) or other tumor-related genes in colorectal cancer (CRC) cells. Required to maintain a transcriptionally repressive state of genes in undifferentiated embryonic stem cells (ESCs). In ESCs, in collaboration with TRIM28, is also required for H3K9me3 and silencing of endogenous and introduced retroviruses in a DNA-methylation independent-pathway. Associates at promoter regions of tumor suppressor genes (TSGs) leading to their gene silencing. The SETDB1-TRIM28-ZNF274 complex may play a role in recruiting ATRX to the 3'-exons of zinc-finger coding genes with atypical chromatin signatures to establish or maintain/protect H3K9me3 at these transcriptionally active regions.
-
基因功能参考文献:
- Study in hepatocellular carcinoma (HCC) cells proves that SETDB1 promotes the proliferation and migration of cells by forming SETDB1-Tiam1 compounds. SETDB1-Tiam1 compounds were involved in a novel pathway, which regulated epigenetic modification of gene expression in HCC patients samples. PMID: 29739365
- SETDB1 loss results in decompaction of the Xi chromosome partly through reactivation of an enhancer in the IL1RAPL1 gene. PMID: 30103804
- SETDB1 knockdown might suppress breast cancer progression at least partly by miR-381-3p-related regulation, providing a novel prospect in breast cancer therapy. PMID: 30309377
- Enhancer of Zeste Homolog 2 (EZH2), SET domain, bifurcated 1 protein (SETDB1), lysine-specific histone demethylase 1 (LSD1), histone H3 methylation (H3K9me3 and H3K27me3) expression are altered in colorectal cancer (CRC) and may play a role in colorectal carcinogenesis. PMID: 30105513
- Increased expression of SETDB1 may predict poor overall survival. PMID: 29901162
- SETDB1 is enriched at H3K9me3 regions and K9me3/K14ac is enriched at SETDB1 binding sites overlapping with LINE elements, suggesting that recruitment of the SETDB1 complex to K14ac/K9me regions has a role in silencing of active genomic regions. PMID: 29234025
- these data identify a critical functional role for ATF7IP in heterochromatin formation by regulating SETDB1 abundance in the nucleus. PMID: 27732843
- SETDB1 triggers silencing of retrotransposons to inhibit the interferon response in acute myeloid leukemia cells. PMID: 28887438
- analysis of a 1q21.3 deletion encompassing SETDB1 that provides further support for the role of chromatin modifiers in the etiology of autism spectrum disorder PMID: 27119313
- SETDB1 protein expression was significantly associated with poor survival and was related to TNM stage. PMID: 28913972
- SETDB1, a major histone H3K9 methyltransferase is monoubiquitinated at the evolutionarily conserved lysine-867 in its SET-Insertion domain. This ubiquitination is directly catalyzed by UBE2E family of E2 enzymes in an E3-independent manner while the conjugated-ubiquitin (Ub) is protected from active deubiquitination. PMID: 27237050
- These results suggest that the ubiquitination of SETDB1 at lysine 867 controls the expression of its target gene by activating its H3K9 methyltransferase activity. PMID: 27798683
- s observed several complementary mechanisms contributing to the upregulation of SETDB1 in HCC cells. Besides copy number gains at the SETDB1 gene locus at chromosome 1q21 enhanced SETDB1 transcription mediated by the transcription factor SP1 could be detected. PMID: 27164857
- BRCA1 and SETDB1 stand out as the most significant prognostic markers in this group of patients PMID: 26542178
- These results suggest that SETDB1- mediated FosB expression is a common molecular phenomenon, and might be a novel pathway responsible for the increase in cell proliferation that frequently occurs during anticancer drug therapy. PMID: 26949019
- SETDB1 mutations are associated with malignant pleural mesotheliomas. PMID: 26824986
- Upon HBV infection, cellular mechanisms involving SETDB1-mediated H3K9me3 and HP1 induce silencing of HBV cccDNA transcription through modulation of chromatin structure. PMID: 26143443
- The SETDB1 protein was closely associated with the prognosis of prostate cancer (PCa). Bioinformatics suggested that SETDB1 might promote PCa bone metastasis through the WNT pathway. In conclusion, SETDB1 might be associated with the development of bone metastases from PCa PMID: 26846621
- results indicate that ATF7IP does not directly modulate SETDB1 catalytic activity, suggesting alternate roles, such as affecting cellular localization or mediating interaction with additional binding partners. PMID: 26813693
- SETDB1 is an oncogene that is frequently up-regulated in human HCCs; the multiplicity of SETDB1 activating mechanisms at the chromosomal, transcriptional, and posttranscriptional levels together facilitates SETDB1 up-regulation in human HCC PMID: 26481868
- regulates cancer cell growth via methylation of p53 PMID: 26471002
- SETDB1, localized in the nucleus, might undergo degradation by the proteasome and be exported to the cytosol, resulting in its detection mainly in the cytosol. PMID: 26296461
- Exogenous expression of MyoD reversed transcriptional repression of MyoD promoter-driven lucif-erase reporter by Setdb1 shRNA and rescued myogenic differentiation of C2C12 myoblast cells depleted of endogenous Setdb1. PMID: 25715926
- s demonstrate that a KMT1E-containing complex directly interacts with the FcgammaRIIb promoter and that histone H3 at lysine 9 tri-methylation at this promoter is dependent on Setdb1 PMID: 25569264
- MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the Epithelial-mesenchymal transition of breast cancer stem cells by down regulating the STAT3 pathway PMID: 25070049
- Report elevated levels of SETDB1 in non-small lung cancers, associated with neoplasm grading and tumor growth. PMID: 25404354
- This observation suggests that the ZNF274/SETDB1 complex bound to the SNORD116 cluster may protect the Prader-Willi syndrome induced pluripotent cells from DNA demethylation during early development. PMID: 24760766
- Together, our findings defined an essential role for the KMT1E/SMAD2/3 repressor complex in TGFbeta-mediated lung cancer metastasis. PMID: 25477335
- SETDB1 is associated with frequent methylation of the euchromatic p16(INK) (4A) promoter and several prognostic parameters in melanomas PMID: 24673285
- data suggested that SETDB1 is overexpressed in human PCa. Silencing SETDB1 inhibited PCa cell proliferation, migration and invasion PMID: 24556744
- overexpression of SETDB1 or LSD1 had no prognostic impact in patients with melanoma PMID: 24658378
- SETDB1 expression was upregulated in glioma cell lines and in glioma tissues compared to normal brain, being positively correlated with grade and histological malignancy. PMID: 23943221
- SETDB1 is a bona fide oncogene undergoing gene amplification-associated activation in lung cancer. PMID: 23770855
- of ESET plays an essential role in the maintenance of articular cartilage by preventing articular chondrocytes from terminal differentiation and may have implications in joint diseases such as osteoarthritis. PMID: 24056368
- A transgenic Setdb1 model established a link between this gene and behavior. PMID: 23055267
- analysis of a KRAB domain-containing ZNF (ZNF274) that is involved in recruitment of the KAP1 and SETDB1 to specific regions of the human genome PMID: 21170338
- studies establish SETDB1 as an oncogene in melanoma and underscore the role of chromatin factors in regulating tumorigenesis PMID: 21430779
- Contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins; KAP-1, SETDB1, H3-MeK9, and HP1 are enriched at promoter sequences of a euchromatic gene silenced by the KRAB-KAP-1 repression system PMID: 11959841
- mAM/hAM facilitates conversion of H3-K9 dimethyl to trimethyl by ESET/SETDB1 PMID: 14536086
- Data show that the methyl-CpG binding protein MBD1 forms a stable complex with histone H3-K9 methylase SETDB1. PMID: 15327775
- These data suggest that MBD1.MCAF1.SETDB1 complex facilitates the formation of heterochromatic domains, emphasizing the role of MCAF/AM family proteins in epigenetic control, and describe a new family member, MCAF2 (ATF7IP2). PMID: 15691849
- histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells PMID: 16682412
- modulation of gene silencing mechanisms, through regulation of the ESET gene is important to neuronal survival and, as such, may be a promising treatment in Huntington's disease patients PMID: 17142323
- Akt/PKB interacts with the histone H3 methyltransferase SETDB1 and coordinates to silence gene expression. PMID: 17577629
- SETDB1 can specifically methylate HIV-1 Tat preferentially at lysine 51. PMID: 18498648
显示更多
收起更多
-
亚细胞定位:Nucleus. Cytoplasm. Chromosome.
-
蛋白家族:Class V-like SAM-binding methyltransferase superfamily, Histone-lysine methyltransferase family, Suvar3-9 subfamily
-
组织特异性:Widely expressed. High expression in testis.
-
数据库链接:
HGNC: 10761
OMIM: 604396
KEGG: hsa:9869
STRING: 9606.ENSP00000271640
UniGene: Hs.643565
Most popular with customers
-
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Myosin regulatory light chain 12A (MYL12A) (Active)
Express system: E.coli
Species: Homo sapiens (Human)
-
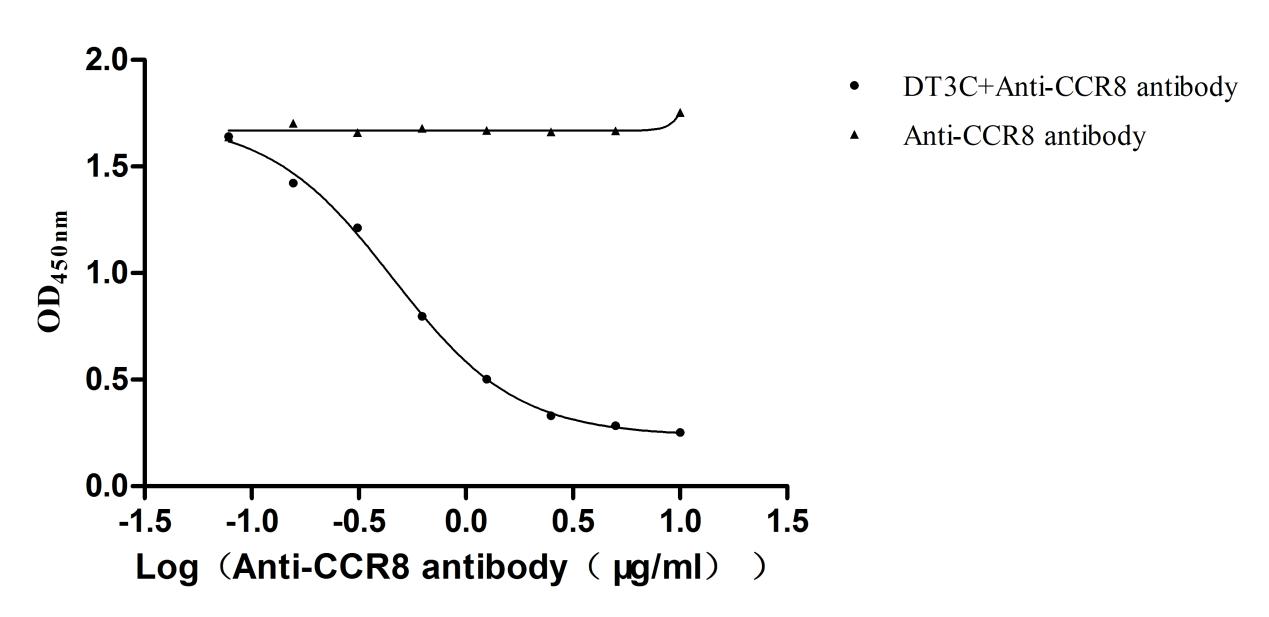
Recombinant DT3C (Diphtheria toxin & spg 3C domain) for Antibody Internalization Assay (Active)
Express system: E.coli
Species: N/A