Recombinant Mouse RNA-binding protein FUS (Fus)
-
中文名称:小鼠Fus重组蛋白
-
货号:CSB-YP009069MO
-
规格:
-
来源:Yeast
-
其他:
-
中文名称:小鼠Fus重组蛋白
-
货号:CSB-EP009069MO
-
规格:
-
来源:E.coli
-
其他:
-
中文名称:小鼠Fus重组蛋白
-
货号:CSB-EP009069MO-B
-
规格:
-
来源:E.coli
-
共轭:Avi-tag Biotinylated
E. coli biotin ligase (BirA) is highly specific in covalently attaching biotin to the 15 amino acid AviTag peptide. This recombinant protein was biotinylated in vivo by AviTag-BirA technology, which method is BriA catalyzes amide linkage between the biotin and the specific lysine of the AviTag.
-
其他:
-
中文名称:小鼠Fus重组蛋白
-
货号:CSB-BP009069MO
-
规格:
-
来源:Baculovirus
-
其他:
-
中文名称:小鼠Fus重组蛋白
-
货号:CSB-MP009069MO
-
规格:
-
来源:Mammalian cell
-
其他:
产品详情
-
纯度:>85% (SDS-PAGE)
-
基因名:Fus
-
Uniprot No.:
-
别名:FusRNA-binding protein FUS; Protein pigpen
-
种属:Mus musculus (Mouse)
-
蛋白长度:Full length protein
-
表达区域:1-518
-
氨基酸序列MASNDYTQQA TQSYGAYPTQ PGQGYSQQSS QPYGQQSYSG YGQSADTSGY GQSSYGSSYG QTQNTGYGTQ SAPQGYGSTG GYGSSQSSQS SYGQQSSYPG YGQQPAPSST SGSYGGSSQS SSYGQPQSGG YGQQSGYGGQ QQSYGQQQSS YNPPQGYGQQ NQYNSSSGGG GGGGGGNYGQ DQSSMSGGGG GGGYGNQDQS GGGGGGYGGG QQDRGGRGRG GGGGYNRSSG GYEPRGRGGG RGGRGGMGGS DRGGFNKFGG PRDQGSRHDS EQDNSDNNTI FVQGLGENVT IESVADYFKQ IGIIKTNKKT GQPMINLYTD RETGKLKGEA TVSFDDPPSA KAAIDWFDGK EFSGNPIKVS FATRRADFNR GGGNGRGGRG RGGPMGRGGY GGGGSGGGGR GGFPSGGGGG GGQQRAGDWK CPNPTCENMN FSWRNECNQC KAPKPDGPGG GPGGSHMGGN YGDDRRGRGG YDRGGYRGRG GDRGGFRGGR GGGDRGGFGP GKMDSRGEHR QDRRERPY
-
蛋白标签:Tag type will be determined during the manufacturing process.
The tag type will be determined during production process. If you have specified tag type, please tell us and we will develop the specified tag preferentially. -
产品提供形式:Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
复溶:We recommend that this vial be briefly centrifuged prior to opening to bring the contents to the bottom. Please reconstitute protein in deionized sterile water to a concentration of 0.1-1.0 mg/mL.We recommend to add 5-50% of glycerol (final concentration) and aliquot for long-term storage at -20℃/-80℃. Our default final concentration of glycerol is 50%. Customers could use it as reference.
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet :Please contact us to get it.
靶点详情
-
功能:DNA/RNA-binding protein that plays a role in various cellular processes such as transcription regulation, RNA splicing, RNA transport, DNA repair and damage response. Binds to nascent pre-mRNAs and acts as a molecular mediator between RNA polymerase II and U1 small nuclear ribonucleoprotein thereby coupling transcription and splicing. Binds also its own pre-mRNA and autoregulates its expression; this autoregulation mechanism is mediated by non-sense-mediated decay. Plays a role in DNA repair mechanisms by promoting D-loop formation and homologous recombination during DNA double-strand break repair. In neuronal cells, plays crucial roles in dendritic spine formation and stability, RNA transport, mRNA stability and synaptic homeostasis.
-
基因功能参考文献:
- a circuitry in which the upregulation of miR-409-3p and miR-495-3p, belonging to a brain-specific miRNA subcluster implicated in several neurodevelopmental disorders, produced the downregulation of Gria2, is reported. PMID: 29430619
- FUS regulates circular RNA biogenesis by binding the introns flanking the back-splicing junctions and that this control can be reproduced with artificial constructs. PMID: 28358055
- FUS and the ELAV-like proteins ELAVL4 and ELAVL1 control SynGAP mRNA stability in a 3'UTR length-dependent manner, resulting in the stable expression of the alternatively spliced SynGAP isoform alpha2. PMID: 28954225
- These two proteins were up-regulated in both HD and FUS/TLS heterozygote mice. PMID: 27739513
- Study established that Fus1 KO mice suffer from the age-related hearing loss (ARHL) of strial origin, making this model a valuable tool for studying mitochondrial/oxidative mechanisms of age-related hearing decline. The model describes the phenotype of premature hearing loss of strial etiology based on Fus1 loss-mediated mitochondrial dysfunction, and identify the target cells and tissues in the inner ear. PMID: 28135838
- s found that FUS, EWS and TAF15 expression is differentially regulated during brain development, both in time and in space. In particular, this study identifies a fine-tuned regulation of FUS and EWS during neuronal differentiation. PMID: 28453628
- Study characterizes a heterozygous knock-in mouse model of ALS and demonstrates that mutations in FUS result in a toxic gain of function leading to motor neuron disease through cell autonomous and non-cell autonomous mechanisms; shows that mutant FUS triggers toxic events in both motor neurons and neighboring cells to elicit motor neuron disease. PMID: 28243725
- We showed that Fus1KO mice develop multiple early aging signs including lordokyphosis, lack of vigor, inability to accumulate fat, reduced ability to tolerate stress, and premature death. PMID: 28351997
- FUS-induced reductions to ER-mitochondria associations and are linked to activation of glycogen synthase kinase-3beta (GSK-3beta), a kinase already strongly associated with ALS/FTD. PMID: 27418313
- our findings indicate that cytoplasmic FUS mislocalization not only leads to nuclear loss of function, but also triggers motor neuron death through a toxic gain of function within motor neurons. PMID: 26951610
- The data of this study support the notion that expression of cytoplasmically mislocalized FUS with compromised RNA-binding capacity causes particularly prominent and harmful FUS pathology in the mouse nervous system. PMID: 25991062
- These results highlight the pivotal role of FUS in regulating GluA1 mRNA stability, post-synaptic function and fronto-temporal lobar degeneration-like animal behaviors. PMID: 25968143
- these studies establish potentially converging disease mechanisms in amyotrophic lateral sclerosis (ALS) and spinal muscular atrophy, with ALS-causative mutants acquiring properties representing both gain and loss of function. PMID: 25625564
- FUS/TLS depletion causes phenotypes possibly related to neuropsychiatric and neurodegenerative conditions, but distinct from ALS and ET, together with specific alterations in RNA metabolisms. PMID: 25907258
- It is associated with amyotrophic lateral sclerosis and its mutation causes accumulation of fus positive stress granules in neurons. PMID: 25216585
- Study provides evidence for loss of PRMT1 function as a consequence of cytoplasmic accumulation of FUS in the pathogenesis of amyotrophic lateral sclerosis, including changes in the histone code regulating gene transcription. PMID: 25274782
- our study provided evidence that a multistep process of FUS aggregation in the cell cytoplasm includes RNA-dependent and RNA-independent mechanisms. PMID: 24842888
- Activation of metabotropic glutamate receptors 1/5 in neocortical slices and isolated synaptoneurosomes increases endogenous mouse FUS and FUS(WT) protein levels but decreases the FUS(R521G) protein PMID: 25324524
- Study shows that neuronal aggregates formed by mutant FUS protein may aberrantly sequester survival motor neuron protein (SMN) and concomitantly cause a reduction of SMN levels in the axon, leading to axonal defects. PMID: 23681068
- FUS is a coregulator of MITF activity and provide new insights into how the RANKL/p38 MAPK signaling nexus controls gene expression in osteoclasts. PMID: 24257758
- Fus directs translation of localized mRNAs in APC-RNP granules. PMID: 24297750
- Characterization of a previously unknown radioprotective function of the mitochondrial tumor suppressor Fus1. PMID: 23788044
- Depletion of neutrophils eliminates the enhanced antibacterial defenses of the Fus1(-/-) mice, suggesting that ultimately it is the enhanced immune cell recruitment that mediates the increased resistance of Fus1(-/-) mice to A. baumannii pneumonia. PMID: 24042119
- Cells with reduced expression of FUS exhibit a loss of cell viability in response to sorbitol, indicating a prosurvival role for endogenous FUS in the cellular response to hyperosmolar stress. PMID: 23625794
- By using a well-established mouse model of Spinal bulbar muscular atrophy, our analysis of primary motor neuron cultures, spinal cords, and microdissected motor neurons show no evidence for FUS dysregulation. PMID: 23062703
- Study reports conserved neuronal RNA targets and networks that are regulated by FUS and that FUS regulates splicing of genes coding for RNA-binding proteins by binding to their highly conserved introns. PMID: 23389473
- PRMT1-mediated arginine methylation could be implicated in the nucleus-cytoplasmic shuttling of FUS/TLS. PMID: 23152885
- FUS regulates alternative splicing events and transcriptions in a position-dependent manner. PMID: 22829983
- We did not observe a significant overlap in the RNA binding sites or the exons regulated by FUS and TDP-43. Nevertheless, we found that both proteins regulate genes that function in neuronal development. PMID: 22934129
- Tau mRNA is identified as a physiological splicing target of FUS. PMID: 22710833
- This review highlights recent advances in understanding the functions of FUS and discusses findings from FUS animal models that provide several key insights into understanding the molecular mechanisms that might contribute to ALS pathogenesis--{REVIEW} PMID: 22342159
- Fus1 establishes its immune- and tumour-suppressive activities via regulation of mitochondrial homeostasis. PMID: 22513871
- Amyotrophic lateral sclerosis mutations in FUS nuclear localization sequence can impair FUS nuclear localization, induce cytoplasmic inclusions and stress granules, and potentially perturb RNA metabolism. PMID: 20674093
- Depletion of PRMT1 diminished the ability of ALS-linked FUS mutants to localize to the cytoplasm. PMID: 21965298
- FUS-CHOP expression in a p53-deficient background is sufficient to initiate liposarcoma in mouse but not in human ASCs, suggesting the need of additional cooperating mutations in hASCs. PMID: 21732477
- Microarray gene expression analysis showed that TLS transcriptional activity is impaired in the absence of PGC-1alpha, and is thus largely dependent on PGC-1alpha PMID: 21338289
- Results demonstrate that amyotrophic lateral sclerosis-linked mutations can variably cause loss of nuclear functions of TLS depending on the degree of impairment in nuclear localization. PMID: 21109527
- Fus as a novel regulator of self-renewal and radioprotection of HSCs and also implicate it in stress-resistance and maintenance of the genomic integrity of HSCs. PMID: 20412831
- Pigpen, a nuclear coiled body component protein, is involved in angiogenesis. PMID: 20148893
- Pigpen encodes a nuclear protein whose expression is developmentally regulated during craniofacial morphogenesis. PMID: 12950080
- TLS participates in mRNA sorting to the dendritic spines induced by mGluR5 activation and regulates spine morphology to stabilize the synaptic structure. PMID: 15797031
- TLS associates with mRNA encoding an actin-related protein and may be involved in actin reorganization in spines PMID: 16317045
- the sequestration of TLS to polyQ aggregates may play a role in diverse pathological changes in the brains of patients with polyQ diseases. PMID: 18167354
- transformation and deregulation of Cdk1 by TLS-ERG require an intact ets DNA-binding domain within the fusion protein PMID: 18505930
- The results identify FUS as a new target of the ATM-signalling pathway and strengthen the notion that FUS regulates genome stability. PMID: 18620545
- IGF1 is a common target gene of Ewing's sarcoma fusion proteins EWS-FLI-1, EWS-ERG and FUS-ERG in mesenchymal progenitor cells PMID: 18648544
- In the present study, myosin VI (Myo6) protein is for the first time shown to interact with Tls in the mouse brain. PMID: 19558455
显示更多
收起更多
-
亚细胞定位:Nucleus.
-
蛋白家族:RRM TET family
-
数据库链接:
KEGG: mmu:233908
STRING: 10090.ENSMUSP00000101858
UniGene: Mm.277680
Most popular with customers
-
Recombinant Rat Intestinal-type alkaline phosphatase 1 (Alpi) (Active)
Express system: Mammalian cell
Species: Rattus norvegicus (Rat)
-
Recombinant Human Claudin-6 (CLDN6)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Interleukin-17A (IL17A) (T26A) (Active)
Express system: Baculovirus
Species: Homo sapiens (Human)
-
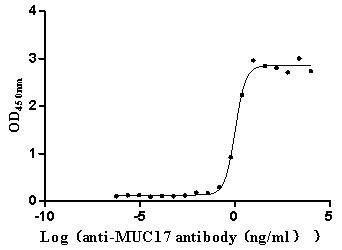
Recombinant Human Mucin-17 (MUC17), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
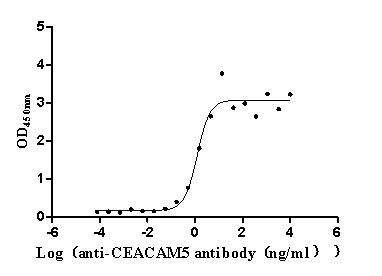
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
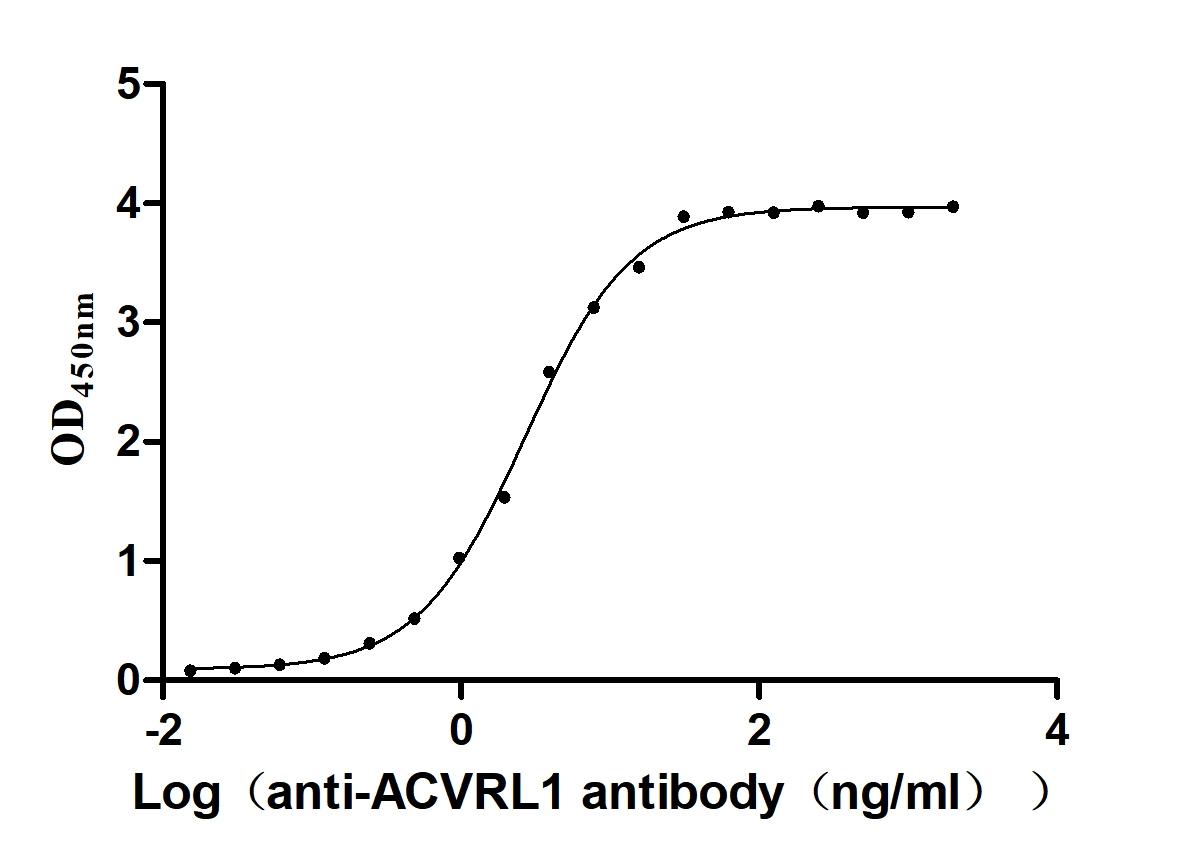
Recombinant Human Serine/threonine-protein kinase receptor R3 (ACVRL1), partial (Active)
Express system: Baculovirus
Species: Homo sapiens (Human)
-
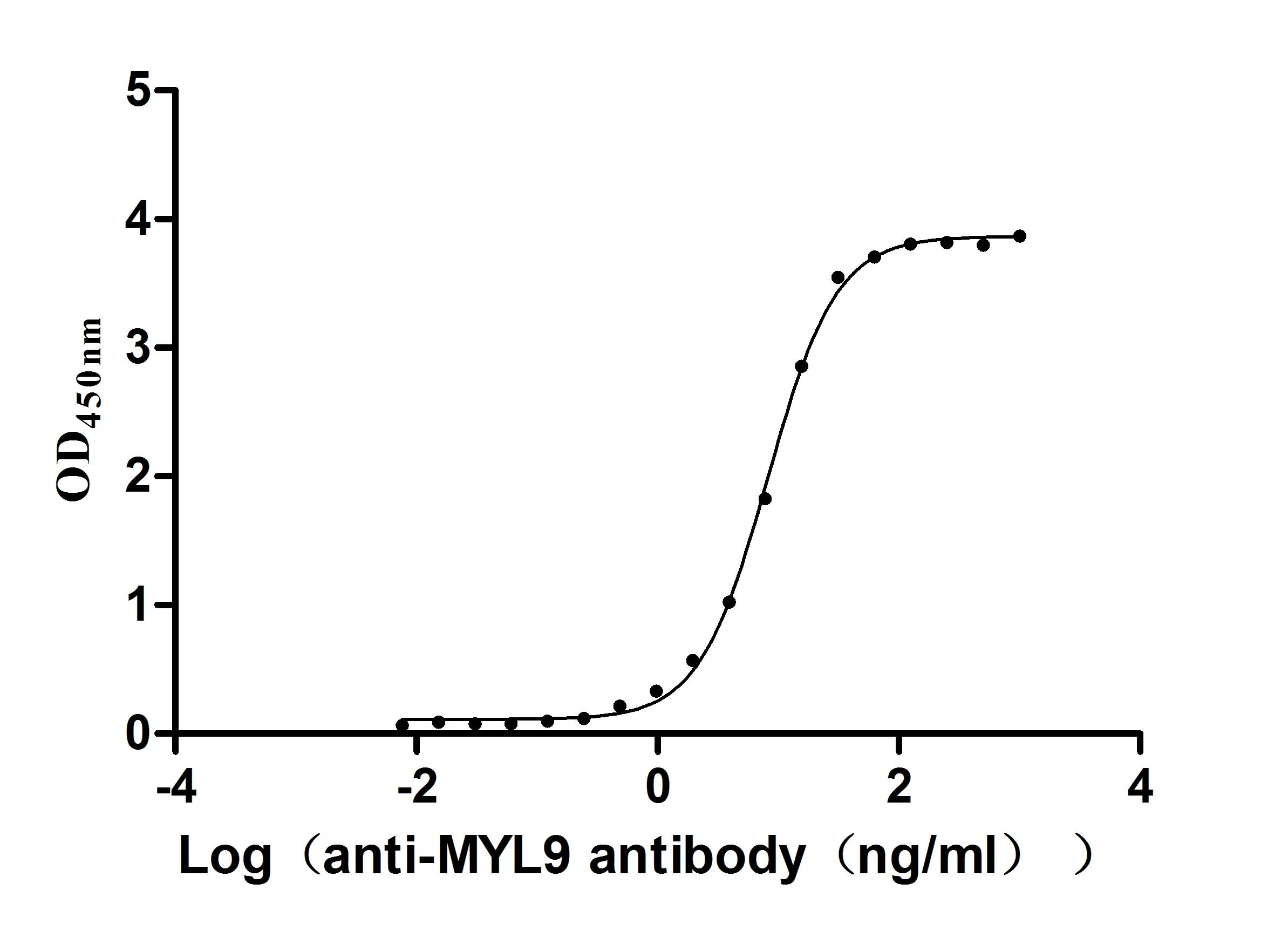
Recombinant Human Myosin regulatory light chain 12B(MYL12B) (Active)
Express system: E.coli
Species: Homo sapiens (Human)
-



-AC1.jpg)
-AC1.jpg)