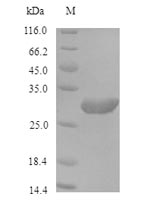
Recombinant Saccharomyces cerevisiae ProlifeRating cell nuclear antigen (POL30)
-
中文名称:酿酒酵母POL30重组蛋白
-
货号:CSB-YP017621SVG
-
规格:¥2616
-
图片:
-
其他:
产品详情
-
纯度:Greater than 90% as determined by SDS-PAGE.
-
基因名:POL30
-
Uniprot No.:
-
别名:POL30; YBR088C; YBR0811; Proliferating cell nuclear antigen; PCNA
-
种属:Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast)
-
蛋白长度:Full Length
-
来源:Yeast
-
分子量:30.9kDa
-
表达区域:1-258aa
-
氨基酸序列MLEAKFEEASLFKRIIDGFKDCVQLVNFQCKEDGIIAQAVDDSRVLLVSLEIGVEAFQEYRCDHPVTLGMDLTSLSKILRCGNNTDTLTLIADNTPDSIILLFEDTKKDRIAEYSLKLMDIDADFLKIEELQYDSTLSLPSSEFSKIVRDLSQLSDSINIMITKETIKFVADGDIGSGSVIIKPFVDMEHPETSIKLEMDQPVDLTFGAKYLLDIIKGSSLSDRVGIRLSSEAPALFQFDLKSGFLQFFLAPKFNDEE
Note: The complete sequence including tag sequence, target protein sequence and linker sequence could be provided upon request. -
蛋白标签:N-terminal 6xHis-tagged
-
产品提供形式:Liquid or Lyophilized powder
Note: We will preferentially ship the format that we have in stock, however, if you have any special requirement for the format, please remark your requirement when placing the order, we will prepare according to your demand. -
缓冲液:Tris-based buffer,50% glycerol
-
储存条件:Store at -20°C/-80°C upon receipt, aliquoting is necessary for mutiple use. Avoid repeated freeze-thaw cycles.
-
保质期:The shelf life is related to many factors, storage state, buffer ingredients, storage temperature and the stability of the protein itself.
Generally, the shelf life of liquid form is 6 months at -20°C/-80°C. The shelf life of lyophilized form is 12 months at -20°C/-80°C. -
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time may differ from different purchasing way or location, please kindly consult your local distributors for specific delivery time.Note: All of our proteins are default shipped with normal blue ice packs, if you request to ship with dry ice, please communicate with us in advance and extra fees will be charged.
-
注意事项:Repeated freezing and thawing is not recommended. Store working aliquots at 4°C for up to one week.
-
Datasheet & COA:Please contact us to get it.
相关产品
靶点详情
-
功能:This protein is an auxiliary protein of DNA polymerase delta and is involved in the control of eukaryotic DNA replication by increasing the polymerase's processibility during elongation of the leading strand. Involved in DNA repair.
-
基因功能参考文献:
- proliferating cell nuclear antigen (PCNA) unloading activity of Elg1 is important for coordinating DNA replication forks with the process of replication-coupled nucleosome assembly to maintain silencing of HML and HMR through S-phase PMID: 29440488
- found evidence implicating PCNA in the maintenance of the high-order structure and stability of heterochromatin, which indicates a role of DNA replication in heterochromatin maintenance PMID: 27987109
- Srs2 interaction with PCNA allows the helicase activity to unwind fork-blocking CAG/CTG hairpin structures to prevent DNA breaks during DNA replication. Independently of PCNA binding, Srs2 also displaces Rad51 from nascent strands to prevent recombination-dependent repeat expansions and contractions. PMID: 28175398
- the various roles of Elg1 in genome stability maintenance involve the unloading of both modified and unmodified PCNA. PMID: 28108661
- Pro-recombination Role of Srs2 Protein Requires SUMO (Small Ubiquitin-like Modifier) but Is Independent of PCNA (Proliferating Cell Nuclear Antigen) Interaction. PMID: 26861880
- results suggest that Elg1-RLC acts as a general PCNA unloader and is dependent upon DNA ligation during chromosome replication. PMID: 26212319
- Elevated levels of PCNA rescue pds5-1 temperature sensitivity and cohesion defects, but do not rescue pds5-1 mutant cell condensation defects. PMID: 25986377
- s discuss the interplay of POL30 and Srs2 proteins in error-free DNA-damage tolerance in Saccharomyces cerevisiae and show how sumoylated PCNA recruits Srs2 for this mechanism. [Review] PMID: 26041265
- In contrast to the "toolbelt" model, which was demonstrated for bacterial and archaeal sliding clamps, our results suggest a mechanism of sequential switching of partners on the eukaryotic PCNA trimer during DNA replication and repair. PMID: 25228764
- Cells deficient in PCNA unloading exhibit increased chromosome breaks. PMID: 25449133
- Data indicate that the C22Y substitution alters the positions of the alpha-helices lining the central hole of the POL30 (PCNA) ring, whereas the C81R substitution creates a distortion in an extended loop near the PCNA subunit interface. PMID: 23869605
- Elg1-replication factor C-like complex functions in unloading of both unmodified and SUMOylated PCNA during DNA replication. PMID: 23499004
- The increased dynamics of hPCNA relative to scPCNA may allow it to acquire multiple induced conformations upon binding to its substrates enlarging its binding diversity. PMID: 21364740
- show that yeast Elg1 interacts physically and genetically with PCNA, in a manner that depends on PCNA modification, and exhibits preferential affinity for SUMOylated PCNA. PMID: 20571511
- High-copy expression of POL30 (PCNA) suppresses the canavanine mutation rate of all the rad27 alleles, including rad27Delta. PMID: 19596905
- SUMO-modified PCNA functionally cooperates with Srs2 PMID: 15931174
- Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase SRs2p. PMID: 15989970
- Mediates the entry of the flap endonuclease and DNA ligase I into the process of Okazaki fragment joining, and this ordered entry is necessary to prevent CAG repeat tract expansions. PMID: 16079237
- PCNA acts as a scaffold for consecutive protein-protein interactions that allow for the coordination of mismatch repair steps. PMID: 16303135
- An open PCNA clamp in a complex with RFC through fluorescence energy transfer experiments was captured. PMID: 16476998
- Diubiquitinated Mcm10 interacts with PCNA to facilitate an essential step in DNA elongation. PMID: 16782870
- Mgs1 physically associates with PCNA and that Mgs1 helps suppress the RAD6 DNA damage tolerance pathway in the absence of exogenous DNA damage. PMID: 16809783
- role for PCNA in regulating the mutagenic activity of Polzeta separate from its modification at Lys164 PMID: 16957771
- The loaded form of yPCNA may play an important effector role in directing yMutLalpha incision to the discontinuous strand of a nicked heteroduplex. PMID: 17951253
- Mismatch repair-PCNA interactions are important for repairing mismatches formed during meiotic recombination, but play only a relatively minor role in regulating the fidelity of mitotic recombination. PMID: 18245822
- PCNA participates in formation and stability of epigenetically regulated chromatin through a pathway that includes replication factor C, the chromatin assembly factor Asf1p, and the K56-specific acetyltransferase Rtt109p. PMID: 18558650
- PCNA modification by SUMO is controlled by DNA PMID: 18701921
- the balance between HDR and NHEJ pathways is crucially controlled by genes of the RAD6 pathway through modifications of PCNA PMID: 18722556
- While wild-type PCNA stimulates incorporation by Dna polymerase eta opposite an abasic site, the mutant PCNA protein actually inhibits incorporation opposite this DNA lesion. PMID: 19053247
- Data suggest that mono-ubiquitination of PCNA is induced by nucleotide misincorporation by DNA polymerase alpha. PMID: 19279190
- DNA binding commits Saccharomyces cerevisiae replication factor C to ATP hydrolysis, which is followed by proliferating cell nuclear antigen (PCNA) closure and PCNA.DNA release. PMID: 19285992
显示更多
收起更多
-
亚细胞定位:Nucleus.
-
蛋白家族:PCNA family
-
数据库链接:
KEGG: sce:YBR088C
STRING: 4932.YBR088C
Most popular with customers
-
Recombinant Human ICOS ligand (ICOSLG), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Tumor necrosis factor ligand superfamily member 9 (TNFSF9), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Glucagon receptor (GCGR), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
Recombinant Human Claudin-4 (CLDN4)-VLPs (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
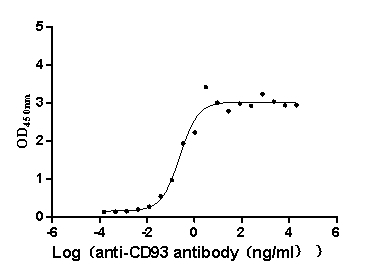
Recombinant Macaca fascicularis CD93 molecule (CD93), partial (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
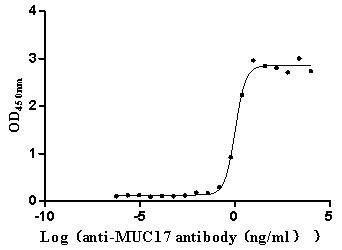
Recombinant Human Mucin-17 (MUC17), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)
-
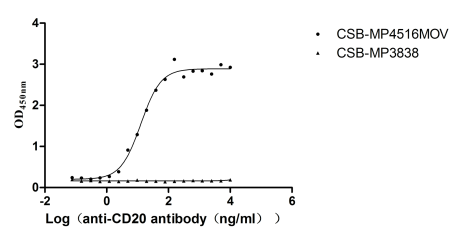
Recombinant Macaca fascicularis Membrane spanning 4-domains A1 (MS4A1)-VLPs (Active)
Express system: Mammalian cell
Species: Macaca fascicularis (Crab-eating macaque) (Cynomolgus monkey)
-
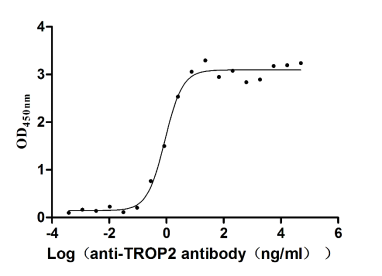
Recombinant Human Tumor-associated calcium signal transducer 2 (TACSTD2), partial (Active)
Express system: Mammalian cell
Species: Homo sapiens (Human)