SUMO1 Antibody
-
货号:CSB-PA042567
-
规格:¥2024
-
图片:
-
其他:
产品详情
-
产品名称:Rabbit anti-Homo sapiens (Human) SUMO1 Polyclonal antibody
-
Uniprot No.:P63165
-
基因名:SUMO1
-
宿主:Rabbit
-
反应种属:Human,Mouse,Rat
-
免疫原:Synthesized peptide derived from N-terminal of Human SUMO-1.
-
免疫原种属:Homo sapiens (Human)
-
克隆类型:Polyclonal
-
纯化方式:The antibody was affinity-purified from rabbit antiserum by affinity-chromatography using epitope-specific immunogen.
-
浓度:It differs from different batches. Please contact us to confirm it.
-
产品提供形式:Liquid
-
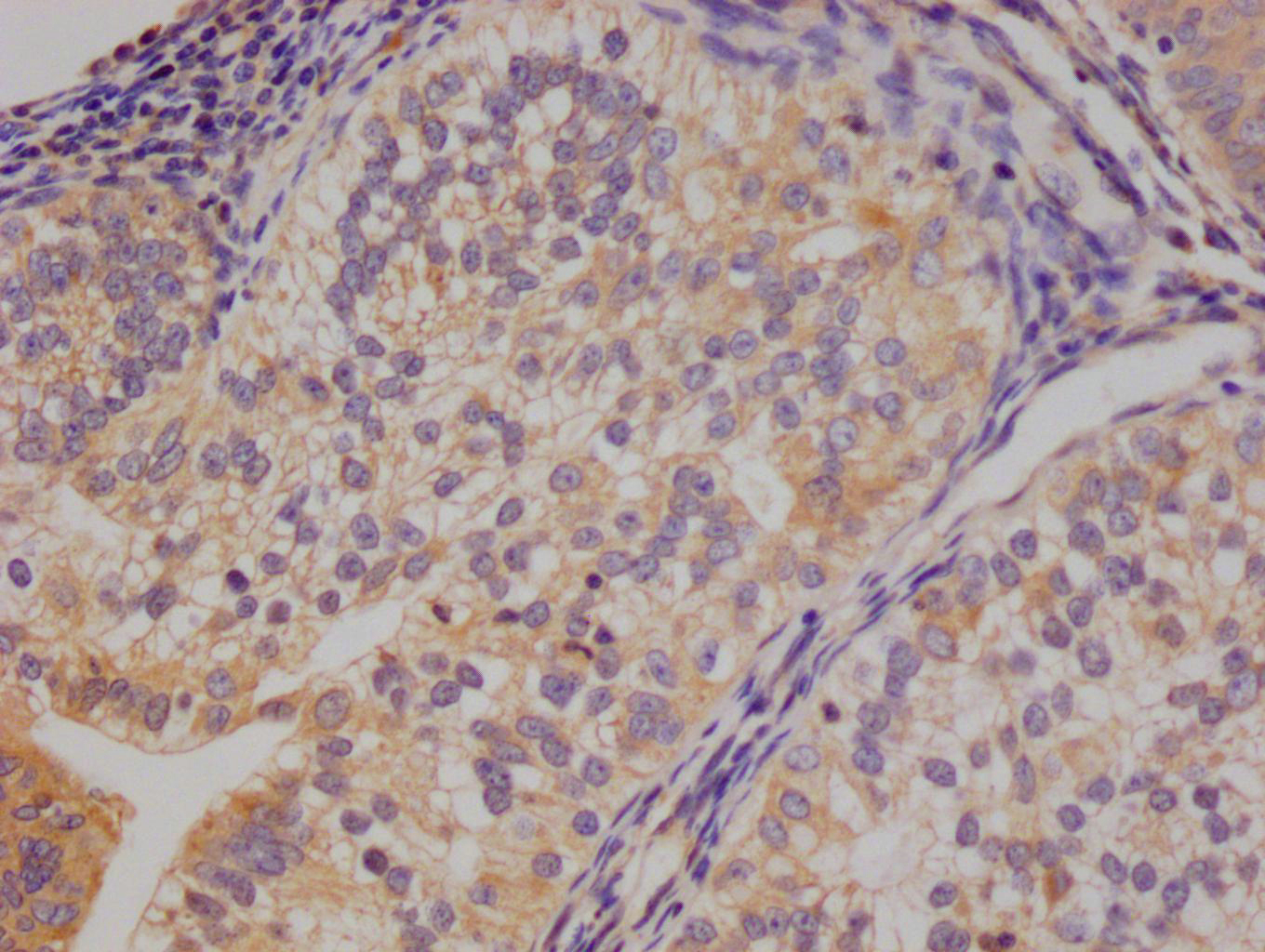
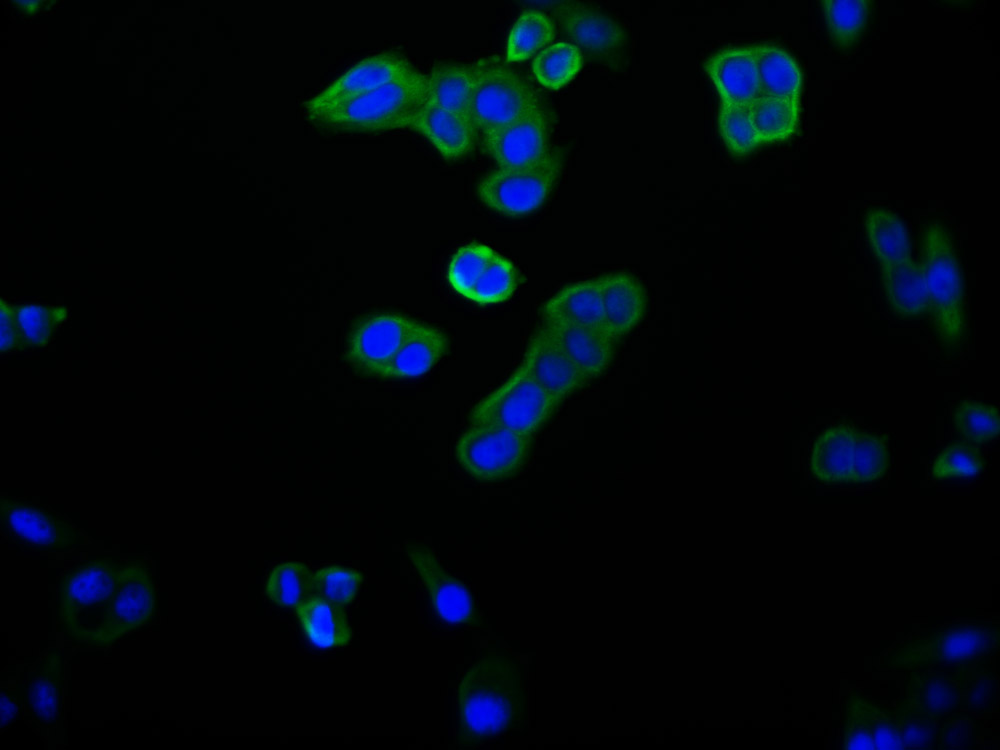
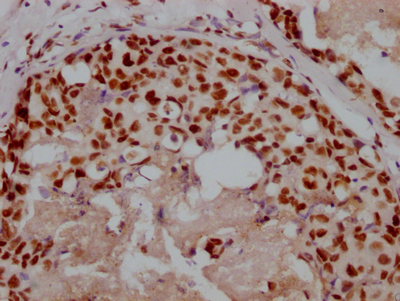
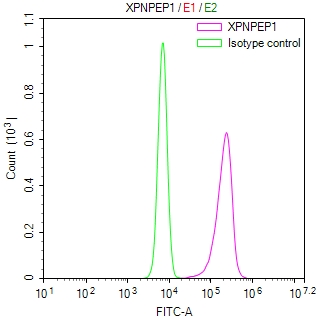
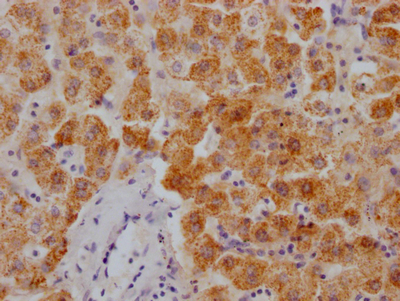
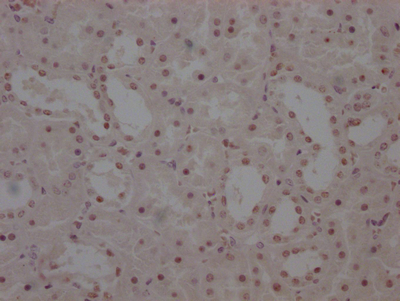
应用范围:ELISA,WB,IHC,IF
-
推荐稀释比:
Application Recommended Dilution WB 1:500-1:3000 IHC 1:50-1:100 IF 1:100-1:500 -
Protocols:
-
储存条件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
货期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
相关产品
靶点详情
-
功能:Ubiquitin-like protein that can be covalently attached to proteins as a monomer or a lysine-linked polymer. Covalent attachment via an isopeptide bond to its substrates requires prior activation by the E1 complex SAE1-SAE2 and linkage to the E2 enzyme UBE2I, and can be promoted by E3 ligases such as PIAS1-4, RANBP2 or CBX4. This post-translational modification on lysine residues of proteins plays a crucial role in a number of cellular processes such as nuclear transport, DNA replication and repair, mitosis and signal transduction. Involved for instance in targeting RANGAP1 to the nuclear pore complex protein RANBP2. Covalently attached to the voltage-gated potassium channel KCNB1; this modulates the gating characteristics of KCNB1. Polymeric SUMO1 chains are also susceptible to polyubiquitination which functions as a signal for proteasomal degradation of modified proteins. May also regulate a network of genes involved in palate development. Covalently attached to ZFHX3.
-
基因功能参考文献:
- SUMO1 expression results in a gain of PKR activity by increasing its activation. SUMO1 was able to activate PKR and eIF-2alpha in the absence of viral infection. PMID: 29352251
- SPOP inhibits hepatocellular carcinoma (HCC) cell metastasis via ubiquitin-dependent SUMO1/SENP7 proteolysis and may thus serve as a new opinion for the prevention of HCC metastasis. PMID: 29777712
- Study found a significant difference in the expression of Cx43 and SUMO1 between cancer stem cells and non-cancer stem cells in liver cancer. By the co-expression of Cx43 and SUMO1 in cancer stem cells, the gap junction intercellular communication of liver cancer stem cells was obviously improved. PMID: 29393359
- Two SUMO modification sites existed in dopamine receptor D1, the phosphorylation of which, due to SUMO modification, can interact with PP2A, leading to the inhibition of D1 de-phosphorylation and normal function. PMID: 28770955
- These results suggest that SUMO1 contributes to hepatocellular carcinoma progression by promoting p65 nuclear translocation PMID: 26993772
- In summary, our study revealed a negative regulation of the UPR transducer ATF6 through post-translational SUMOylation. The information from this study will not only increase our understanding of the fine-tuning regulation of the UPR signaling but will also be informative to the modulation of the UPR for therapeutic benefits. PMID: 29061306
- Molecular dynamics simulations showed that binding of the beta-grasp domain of SUMO1 induces significant conformational and dynamic changes in SENP1, including widening of the exosite cleft and quenching of nanosecond dynamics in all but a distal region. PMID: 27576863
- Mutational analysis of functional sites showed that both peroxidase and PLA2 active sites were necessary for mutant Prdx6 function, and that Prdx6 phosphorylation (at T177 residue) was essential for optimum PLA2 activity.Mutant Prdx6 at its Sumo1 sites escapes and abates this adverse process by maintaining its integrity and gaining function PMID: 28055018
- SUMO and p21Cip1 regulate the transit of proteins through the nucleolus; disruption of nucleolar export by DNA damage induces SUMO and p21Cip1 to act as hub proteins to form a multiprotein complex in the nucleolus. PMID: 28582471
- This study reveals an essential role of SUMOylated FADD in Drp1- and caspase-10-dependent necrosis. PMID: 27799292
- SUMO-1 gene silencing inhibits proliferation and promotes apoptosis of human gastric cancer cells. PMID: 28222440
- the critical role of Cys52 in maintaining SUMO-1 conformation and function PMID: 27195426
- Findings suggest SUMO-1 protein and PGE2 receptor subtype 4 (EP4) as two potential targets for new therapeutic or prevention strategies for endometrial cancers. PMID: 27230680
- This study demonstrated that the rs12472035 polymorphism of SUMO1 was significantly associated with an increased risk of AD in male group. PMID: 27084229
- FOXP2 can be modified with all three human SUMO proteins and that PIAS1 promotes this process. PMID: 26867680
- Ang II-induced upregulation of ATF3 and SUMO1 in vitro and in vivo was blocked by Ang II type I receptor antagonist olmesartan. Moreover, Ang II induced ATF3 SUMOylation at lysine 42, which is SUMO1 dependent. PMID: 26850942
- Data show that mutation of key residues in the binding site abolishes binding and that small ubiquitin-like modifier 1 (SUMO1) can simultaneously and non-cooperatively bind both the ZZ domain and a canonical SIM motif of CREB-binding protein (CBP/p300). PMID: 27129204
- Roles for SUMO in pre-mRNA processing PMID: 26563097
- SUMOylation at specific sites on PXR protein are involved in enhancement of transcription function of this receptor. PMID: 26549688
- High DAP1 expression is associated with a 4-fold increase in the risk of lymph node metastases in squamous cell carcinoma of the oral cavity. PMID: 26400283
- Knockdown of SUMO1 using specific siRNA influenced the accumulation of lipid droplets and reduced HCV replication. PMID: 26449956
- The LKB1 K178R SUMO mutant had defective AMPK signaling and mitochondrial function, inducing death in energy-deprived cells. PMID: 26212320
- Data suggest that small ubiquitin-related modifier protein SUMO1 modification of the promyelocytic leukemia protein (PML) RING domain promotes SUMO2 conjugation to Lys160. PMID: 26060329
- Data identify PDGFRbeta as the hub gene in both inflammatory (IBC) and non-inflammatory breast neoplasm (non-IBC) and SUMO1 and COL1A1 the respective key genes for IBC and non-IBC suggesting they might play important role in the pathogenesis of the neoplasm. PMID: 26403314
- PML IV/ARF interaction enhances p53 SUMO-1 conjugation, activation, and senescence. PMID: 26578773
- Data indicate that small ubiquitin-like modifiers SUMO1, SUMO2, or SUMO3 were found in nuclear speckles. PMID: 26223657
- These results confirm that the SUMO machinery is involved in TRIM5alpha-mediated retroviral restriction, and demonstrate that TRIM5alpha is a SUMO 1 and SUMO 2 substrate. PMID: 25880753
- Under cell-free in vitro conditions, p35 is covalently modified by SUMO1. Sumoylation is a likely candidate mechanism for the rapid modulation of p35/Cdk5 activity in physiological situations as well as in disease. PMID: 25391294
- SUMO1 modification stabilizes CDK6 protein and drives the cell cycle and glioblastoma progression. PMID: 24953629
- SUMO1 accelerates the accumulation of autophagic vacuoles and promotes Abeta production. PMID: 25484073
- Expression of SUMO1/2/3 is dramatically enhanced by interferons through an miRNA-based mechanism involving the Lin28/let-7. PMID: 24942926
- SUMO-1 modification may affect the transcriptional activity of EGFR PMID: 26244656
- The crystal structures of SUMO1 bound to unphosphorylated and tetraphosphorylated PML-SUMO-interacting motifs peptides indicate that three phosphoserines directly contact specific positively charged residues of SUMO1. PMID: 25497731
- The expression of SUMO-1 in OLP was similar to normal mucosa and inflammatory fibrous hyperplasia, suggesting that alterations of this protein occur at later stages of carcinogenesis PMID: 23350884
- reveals an unexpected role of SUMO-1 and SAFB in the stimulatory coupling of promoter binding, transcription initiation and RNA processing PMID: 25800734
- MDM2 and SUMO-1 proteins are up-regulated in actinic cheilitis and lip cancer. PMID: 25241153
- Identify DRP1, RanGAP1 and topoisomerase IIa as targets of SUMO1 in human ejaculated sperm, giving more insights on the role of SUMO1-ylation in sperm morphology and motility. PMID: 25118297
- findings provide empirical evidence that SUMO1 genetic polymorphisms might be strongly involved in the etiology of NSCL/P, especially for rs12470401 T>C, rs16838917 A>G, rs12470529 A>G, and rs7572505 A>G polymorphisms. PMID: 25111678
- Overexpression of SUMO-1 decreased phosphorylated GSK-3beta at Ser-9. Mutagenesis of tau at K340R or inhibition of tau SUMOylation by ginkgolic acid abolishes the effect of SUMO-1. PMID: 25378699
- The mutation of the sumoylation site (Lys137) of PTBP2 markedly inhibited its modification by SUMO1. PMID: 24286314
- SUMO potentiates the inhibition of protein synthesis induced by PKR in response to dsRNA. PMID: 25074923
- These results show that DAP1 is a key regulator, of the induction of apoptosis and reduction of autophagy by subtilase cytotoxin (SubAB). PMID: 25183729
- identified 295 SUMO1 and 167 SUMO2 sites on endogenous substrates of HeLa cells PMID: 25114211
- Modification of TDG by small ubiquitin-like modifier (SUMO) proteins weakens its binding to abasic DNA. PMID: 24753249
- SUMO-1 modification of RARA is a potent mechanism for balancing proliferation and differentiation by controlling the stability of RARA in cancer cells. PMID: 24819975
- SUMO1P3 was significantly up-regulated in gastric cancer tissues. PMID: 23996296
- SUMO chain-induced dimerization activates RNF4. PMID: 24656128
- The mitosis-dependent dynamic SUMO-1 modification of NuMA might contribute to NuMA-mediated formation and maintenance of mitotic spindle poles during mitosis. PMID: 24309115
- PIASxalpha is a novel SUMO E3 ligase for PTEN, and it positively regulates PTEN protein level in tumor suppression. PMID: 24344134
- SUMO participates in transcriptional repression as both a covalent modification and through non-covalent interactions with E2 and E3 enzymes. PMID: 24174529
显示更多
收起更多
-
相关疾病:Non-syndromic orofacial cleft 10 (OFC10)
-
亚细胞定位:Nucleus membrane. Nucleus speckle. Cytoplasm. Nucleus, PML body. Cell membrane. Nucleus.
-
蛋白家族:Ubiquitin family, SUMO subfamily
-
数据库链接:
HGNC: 12502
OMIM: 601912
KEGG: hsa:7341
STRING: 9606.ENSP00000376076
UniGene: Hs.81424
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IF, FC
Species Reactivity: Human, Mouse, Rat
-
-
-
-
-
-